| 1 |
Biomolecular Sructure and Modeling: Historical Perspective
|
|
1.1 Simulation evolution (3D version) [J. Huang]
1.2 Simulation evolution (2D version) [J. Huang, K. Schulten, E. Tajkhorshid, D. Strahs, P. Kollman, D. Beveridge]
1.3 Cryo-EM view of 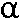 - latrotoxin [H.H. Gan, Y. Ushkaryov] - latrotoxin [H.H. Gan, Y. Ushkaryov]
|
|
| 2 |
Biomolecular Structure and Modeling: Problem and Application Perspective |
|
2.1 Sequence and Structure Data [M. Ding, J. Huang]
2.2 Paracelsus' Janus [H.H. Gan]
2.3 GroEL/GroES chaperonin/co-chaperonin complex [J. Noah]
2.4 Prion protein [J. Noah]
2.5 AIDS drugs [L. Yang]
|
|
| 3 |
Protein Basic |
|
3.1 An amino acid [J. Huang, W. Xu]
3.2 Water clusters [L. Yang, D. Xie, D. Strahs]
3.3 Dipeptide formation [J. Huang]
3.4 Peptide formula [J. Huang, W. Xu]
3.5 Aspartame [W. Xu]
3.6 The amino acid repertoire [J. Huang]
3.7 Amino acids structures [J. Huang, W. Xu]
3.8 Amino acid frequencies [H.H. Gan, J. Noah]
3.9 Fibrous proteins [H.H. Gan, H. Berman]
3.10 Rop [H.H. Gan]
3.11 EF Proteins [H.H. Gan]
3.12 Protein-structure variants [H.H. Gan]
3.13 Gauche and trans orientations [W. Xu]
3.14 Dihedral angle [M. Ding]
3.15 Rotations in polypeptides [J. Huang, W. Xu]
3.16 Amino acids rotamers [J. Huang]
3.17 Ramachandran plots [J. Noah, H.H. Gan]
3.18 Further Study of Ramachandran plots [J. Noah, H.H. Gan]
|
|
| 4 |
Protein Hierarchy |
|
4.1 The 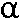 - helix and - helix and
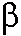 - sheet motifs [R. Sinovcic, J. Huang] - sheet motifs [R. Sinovcic, J. Huang]
4.2 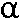 - helical proteins (a) [H.H. Gan] - helical proteins (a) [H.H. Gan]
4.3 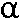 - helical proteins (b) [H.H. Gan] - helical proteins (b) [H.H. Gan]
4.4 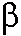 - helical proteins (a) [H.H. Gan] - helical proteins (a) [H.H. Gan]
4.5 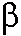 - helical proteins (b) [H.H. Gan] - helical proteins (b) [H.H. Gan]
4.6 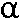 / / 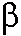 proteins [H.H. Gan]
proteins [H.H. Gan]
4.7 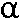 + + 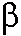 proteins [H.H. Gan]
proteins [H.H. Gan]
4.8 Tomato bushy stunt virus [H.H. Gan]
| |
| 5 |
Nucleic Acids Structure |
|
5.1 The DNA double helix [D. Strahs]
5.2 Nucleic acid components [R. Sinovcic, J. Huang, D. Strahs]
5.3 Watson-Crick base pairing [R. Sinovcic, J. Huang, D. Strahs]
5.4 The polynucleotide chain and labeling [R. Sinovcic, J. Huang]
5.5 Sugar envelope and twist puckers [D. Strahs]
5.6 Sugar pseudorotation cycle [W. Xu, J. Huang]
5.7 Common sugar puckers [D. Strahs]
5.8 Sugar pucker clustering [J. Huang]
5.9 Torsion angle wheel [J. Huang, D. Strahs]
5.10 Deoxyadenosine adiabatic map [D. Strahs]
5.11 Base-pair coordinate system [J. Huang]
5.12 Base-pair step and base pair parameters [D. Strahs]
5.13 Model A, B, and Z-DNA [D. Strahs]
5.14 Model A, B, and Z-DNA, stereo side [D. Strahs]
5.15 Model A, B, and Z-DNA, stereo top [D. Strahs]
|
|
| 6 |
Topics in Nucleic Acids Structure |
|
6.1 Bending in long DNA [D. Strahs]
6.2 Net DNA bending examples [D. Strahs]
6.3 A-tract DNA dodecamer [D. Strahs]
6.4 Sequence-dependent local DNA hydration [D. Strahs]
6.5 DNA/protein binding motifs [D. Strahs]
6.6 Various hydrogen-bonding schemes [R. Sinovcic, J. Huang, D. Strahs]
6.7 DNA/protein complex with Hoogsteen bp [C. Wolberger, J. Noah, J. Aishima, H.H. Gan]
6.8 Oligonucleotide analogues [L. Yang]
6.9 Various nucleotide-chain folding motifs [D. Strahs]
6.10 RNAs with pseudoknots [D. Strahs]
6.11 Interwound and toroidal supercoiling [D. Beard, J. Huang]
6.12 Nucleosome core particle [D. Beard]
6.13 Schematic view of DNA levels of folding [D. Beard]
6.14 Supercoiling topology and geometry [W. Xu, J. Huang]
6.15 Brownian dynamics snapshots of DNA [J. Huang]
6.16 Site juxtaposition measurements [J. Huang]
6.17 Polynucleosome modeling [D. Beard]
|
|
| 7 |
Theoretical Approaches |
|
7.1 DNA quantum-mechanically derived electrostatic potentials [D. York]
7.2 Enolase active site [W. Yang, H. Liu, Y. Zhang]
7.3 Molecular geometry [W. Xu]
7.4 CHARMM Atom types [J. Huang]
|
|
| 8 |
Force Fields |
|
8.1 Normal modes of a water molecule [W. Xu]
8.2 Computed protein and water spectra [A. Sandu]
8.3 Vibrational modes types [W. Xu]
8.4 Bond-length potentials [L. Yang]
8.5 Bond angles [L. Yang]
8.6 Bond-angle potentials [L. Yang]
8.7 Stretch/bend cross terms [L. Yang]
8.8 Butane torsional orientations [L. Yang]
8.9 Torsion-angle potentials [L. Yang]
8.10 Model compounds for torsional parameterization [L. Yang]
8.11 Wilson angle [L. Yang]
8.12 Van der Waals potentials [L. Yang]
8.13 Coulombic potentials [L. Yang, J. Huang]
|
|
| 9 |
Nonbonded Computations |
|
9.1 CPU Time for Nonbonded Calculations [D. Xie, D. Strahs]
9.2 Cutoff schemes [D. Xie]
9.3 Switch and shift functions [X. Qian]
9.4 Periodic domains [D. Beard]
9.5 Various periodic domains [X. Qian, D. Strahs, L. Yang]
9.6 Space-filling polyhedra [D. Strahs, X. Qian]
9.7 Ewald's trick of Gaussian masking [D. Beard]
9.8 CPU Time for PME vs. Fast Multipole [J. Board]
9.9 Fast multipole schemes [D. Beard]
9.10 Screened Coulomb potential [J. Huang]
9.11 Poisson-Boltzmann Rendering of the 30S Ribosome [A. McCammon, N. Baker]
|
|
| 10 |
Multivariate Minimization |
|
10.1 One-dimensional function [D. Strahs]
10.2 2D Contour curves for quadratic functions [D. Beard]
10.3 3D curves for quadratic functions [D. Beard]
10.4 Sparse Hessians [D. Xie, J. Huang]
10.5 Sparse Hessians, continued [D. Xie, J. Huang]
10.6 Line search minimization [M. Ding]
10.7 Newton's method, simple illustration [K. Arora]
10.8 Newton's method, quadratic example output
10.9 Newton's method, cubic example output
10.10 Minimization paths [Q. Zhang]
10.11 Minimization progress [D. Xie]
|
|
| 11 |
Monte Carlo Techniques |
|
11.1 Lattice structure for simple random number generators
11.2 Lattice structure for linear congruential generators [X. Qian and D. Barash]
11.3 MC computation of 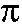 [X. Qian] [X. Qian]
11.4 Boltzmann probabilities [W. Xu, J. Huang]
11.5 MC moves for DNA [J. Huang]
11.6 MC and BD DNA Distributions [J. Huang]
11.7 Bad MC Protocol [J. Huang]
|
|
| 12 |
Molecular Dynamics: Basics |
|
12.1 Sampling methods [J. Huang, D. Strahs, H.H. Gan, L. Yang, S. Broyde, D. Patel, A. McCammon, D. Sept, K. Tai]
12.1 Equilibration [D. Strahs]
12.2 Chaos in MD [J. Huang, X. Qian]
12.3 Butane's end-to-end distance [A. Sandu]
12.4 Butane's end-to-end distance convergence [A. Sandu]
12.5 Energy drift [A. Sandu, J. Huang]
|
|
| 13 |
Molecular Dynamics: Further Topics |
|
13.1 Effective Verlet phase space rotation [W. Xu, J. Huang]
13.2 Verlet resonance for a Morse oscillator [W. Xu, J. Huang]
13.3 Extrapolative vs. Impulse MTS [M. Mandziuk, X. Qian]
13.4 Impulse vs. Extrapolative Force Splitting [E. Barth, J. Huang]
13.5 Resonance from Force Splitting [E. Barth, A. Sandu]
13.6 Harmonic Oscillator Langevin Trajectories [X. Qian]
13.7 BPTI means and variances by Langevin and Newtonian force splitting [A. Sandu, J. Huang]
13.8 LN algorithm
13.9 Manhattan plots for polymerase/DNA [L. Yang]
13.10 Polymerase/DNA geometry [L. Yang]
13.11 BPTI spectral densities [A. Sandu]
13.12 Polymerase/DNA spectral densities [L. Yang]
13.13 Polymerase/DNA geometry [L. Yang]
13.14 Cholesky vs. Chebyshev approaches for the BD random force [D. Beard, J. Huang, M. Ding]
13.15 Implicit and explicit Euler [P. Batcho]
13.16 Verlet and implicit-midpoint energies [M. Mandziuk, G. Zhang]
13.17 Stochastic-path approach snapshots [K. Siva, R. Elber]
|
|
| 14 |
Similarity and Diversity |
|
14.1 Sample drugs [L. Yang]
14.2 Related pairs of drugs [L. Yang]
14.3 Chemical library [M. Ding, L. Yang]
14.4 SVD/refinement performance [D. Xie]
14.5 SVD-based database projection in 2D and 3D [J. Huang, D. Xie]
14.6 Cluster analysis [J. Huang, M. Ding, D. Xie]
14.7 PCA projection in 2D, with similar pairs [J. Huang, D. Xie, S. Singh, E. Fluder]
14.8 PCA projection in 2D, with diverse pairs [J. Huang, D. Xie, S. Singh, E. Fluder]
|
|
| Appendix D Homeworks |
|
D.1 Sample histogram for protein/DNA interaction analysis [D. Strahs]
D.2 Biphenyl [M. Mandziuk]
D.3 Structure for linear congruential generators [D. Barash]
|

 Back to the Main Page of the Book
Back to the Main Page of the Book