In Silico Evidence for DNA Polymerase 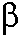 's Substrate-Induced Conformational Change
's Substrate-Induced Conformational Change
Structural information for mammalian DNA polymerase 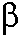 combined with molecular and essential
dynamics studies provide atomistically detailed views of functionally important conformational
rearrangements that occur during DNA repair and replication. Anchors for our study are
available in crystallographic structures of the DNA polymerase
combined with molecular and essential
dynamics studies provide atomistically detailed views of functionally important conformational
rearrangements that occur during DNA repair and replication. Anchors for our study are
available in crystallographic structures of the DNA polymerase 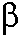 in ``open'' (polymerase bound to gapped
DNA) and ``closed'' (polymerase bound to gapped DNA and substrate, dCTP) forms; these different
states suggest a large-scale conformational change that may help the polymerase choose the correct
nucleotide, and hence monitor DNA synthesis fidelity, through an `induced-fit' mechanism. Our
dynamics simulations of polymerase
in ``open'' (polymerase bound to gapped
DNA) and ``closed'' (polymerase bound to gapped DNA and substrate, dCTP) forms; these different
states suggest a large-scale conformational change that may help the polymerase choose the correct
nucleotide, and hence monitor DNA synthesis fidelity, through an `induced-fit' mechanism. Our
dynamics simulations of polymerase 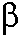 /DNA reveal the large-scale closing motions of the thumb and 8-kDa
subdomains in the presence of the correct substrate -- leading to nearly perfect rearrangement of
residues in the active site for the subsequent chemical step of nucleotidyl transfer -- in contrast to
an opening when the substrate is absent, leading to complete disassembly of the active site residues.
These studies provide in silico evidence for the substrate induced conformational rearrangements
hypothesized experimentally. Further details gleaned from essential dynamics analyses clarify functionally
relevant global motions of the polymerase
polymerase
/DNA reveal the large-scale closing motions of the thumb and 8-kDa
subdomains in the presence of the correct substrate -- leading to nearly perfect rearrangement of
residues in the active site for the subsequent chemical step of nucleotidyl transfer -- in contrast to
an opening when the substrate is absent, leading to complete disassembly of the active site residues.
These studies provide in silico evidence for the substrate induced conformational rearrangements
hypothesized experimentally. Further details gleaned from essential dynamics analyses clarify functionally
relevant global motions of the polymerase
polymerase 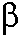 /DNA complex.
/DNA complex.

 Click to go back to the publication list
Click to go back to the publication list
