Search Strategies, Minimization Algorithms, and Molecular Dynamics Simulations
for Exploring Conformational Spaces of Nucleic Acids
We describe recent computational strategies employed in our nucleic acid
programs DUPLEX (in dihedral angle space) and MADPAC (in cartesian space).
Under the first category of Search Strategies, we discuss: i) a build-up
technique for nucleic acids, ii) the use of "soft" constraints to locate
structures that match NMR data or to locate specific hydrogen-bonding patterns,
and iii) a helix generator for single and double-stranded RNA and DNA.
In the second category of Minimization, we describe a rapidly convergent
truncated Newton method specifically adapted for potential energy functions.
Third, in the category of Molecular Dynamics, we introduce a new algorithm
based on the Langevin equation and the implicit Euler Scheme. The implicit
formulation relaxes the severe restriction on the time step size (as in
explicit schemes) and, furthermore, allows incorporation of a "cutoff"
frequency 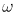 c which
effectively freezes modes of much higher frequency than
c which
effectively freezes modes of much higher frequency than 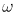 c.
c.

 Click to go back to the publication list
Click to go back to the publication list
Webpage design by Igor Zilberman

 Click to go back to the publication list
Click to go back to the publication list