Beyond Optimization: Simulating the Dynamics of Supercoiled DNA by a Macroscopic
Model
The development and application of macroscopic dynamic models for large
biomolecules is as much of an art as it is a science. Here we discuss scaling
and parameterization issues for a macroscopic B-spline model of supercoiled
DNA in the context of the Langevin framework. We show how scaling the masses
and the damping constant, 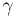 ,
is computationally advantageous (for enhanced sampling) and how, by calibration,
the corresponding physical timestep and damping constant are obtained.
Our timestep estimate for a mass scaled model, roughly 20 ps, is consistent
with a rigorous calibration for the dynamic B-spline model performed
earlier. An interesting finding here is that the B-spline model, though
different in details from a bead model, is essentially equivalent in the
Langevin framework; thus, its computational simplicity can be advantageous.
Another finding is that the optimal
,
is computationally advantageous (for enhanced sampling) and how, by calibration,
the corresponding physical timestep and damping constant are obtained.
Our timestep estimate for a mass scaled model, roughly 20 ps, is consistent
with a rigorous calibration for the dynamic B-spline model performed
earlier. An interesting finding here is that the B-spline model, though
different in details from a bead model, is essentially equivalent in the
Langevin framework; thus, its computational simplicity can be advantageous.
Another finding is that the optimal 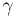 for sampling is smaller (by two orders of magnitude) than the value needed
to reproduce the experimental translational diffusion constant, Dt.
This has been noted previously for proteins and appears to be an important
aspect of modeling macromolecules in view of the sampling problem, as well
as numerical integration. Using the smaller
for sampling is smaller (by two orders of magnitude) than the value needed
to reproduce the experimental translational diffusion constant, Dt.
This has been noted previously for proteins and appears to be an important
aspect of modeling macromolecules in view of the sampling problem, as well
as numerical integration. Using the smaller 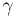 has the effect, in the diffusive regime, of scaling the times computed
in a Langevin trajectory by a corresponding factor. This leads to an effective
timestep in our simulation around 10 ns. Performance of the implicit integrator
used here is also discussed, demonstrating that numerical damping effects
are negligible for this macroscopic model at the
has the effect, in the diffusive regime, of scaling the times computed
in a Langevin trajectory by a corresponding factor. This leads to an effective
timestep in our simulation around 10 ns. Performance of the implicit integrator
used here is also discussed, demonstrating that numerical damping effects
are negligible for this macroscopic model at the 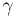 used for maximal sampling. Furthermore, computational gain can be achieved
over explicit integration. Finally, an estimate on the effects of hydrodynamics
on the translational friction constant, fT, is presented, on
the basis of the Kirkwood-Riseman equation. We find that fT
is
reduced by approximately a factor of two for DNA of 1000 base pairs, making
the free-draining limit (i.e., no hydrodynamics) quite reasonable. The
results presented here - scaling, parameterization, and integration in
the context of a dynamic macroscopic model - have general applicability
to simplified models of biopolymers. They also add the needed qualitative
information so that slow dynamic processes involving supercoiled DNA, such
as slithering, can be interpreted in terms of physical timescales from
such macroscopic dynamic simulations.
used for maximal sampling. Furthermore, computational gain can be achieved
over explicit integration. Finally, an estimate on the effects of hydrodynamics
on the translational friction constant, fT, is presented, on
the basis of the Kirkwood-Riseman equation. We find that fT
is
reduced by approximately a factor of two for DNA of 1000 base pairs, making
the free-draining limit (i.e., no hydrodynamics) quite reasonable. The
results presented here - scaling, parameterization, and integration in
the context of a dynamic macroscopic model - have general applicability
to simplified models of biopolymers. They also add the needed qualitative
information so that slow dynamic processes involving supercoiled DNA, such
as slithering, can be interpreted in terms of physical timescales from
such macroscopic dynamic simulations.

 Click to go back to the publication list
Click to go back to the publication list
Webpage design by Igor Zilberman

 Click to go back to the publication list
Click to go back to the publication list