Polymerase 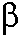 Simulations Reveal That Arg258 Rotation is a Slow
Step Rather Than Large Subdomain Motions Per Se
Simulations Reveal That Arg258 Rotation is a Slow
Step Rather Than Large Subdomain Motions Per Se
The large-scale opening motion of mammalian polymerase
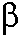 is followed
at atomic resolution by dynamic simulations that link crystal `closed' and
`open' conformations. This conformational change is thought to be key to
the ability of polymerases to choose a correct nucleotide and hence
maintain DNA synthesis fidelity. Corroborating available structural and
kinetic measurements, our studies delineate a sequence of steps after the
chemical DNA extension step: Phe272 ring flip, large thumb movement, and
Arg258 rotation with release of catalytic Mg2+. Significantly, we
identify the last step as slow rather than large subdomain motion
per se. In the closing, these slow rearrangements could be
further hampered if pol
is followed
at atomic resolution by dynamic simulations that link crystal `closed' and
`open' conformations. This conformational change is thought to be key to
the ability of polymerases to choose a correct nucleotide and hence
maintain DNA synthesis fidelity. Corroborating available structural and
kinetic measurements, our studies delineate a sequence of steps after the
chemical DNA extension step: Phe272 ring flip, large thumb movement, and
Arg258 rotation with release of catalytic Mg2+. Significantly, we
identify the last step as slow rather than large subdomain motion
per se. In the closing, these slow rearrangements could be
further hampered if pol 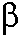 binds to an incorrect nucleotide. Our
results suggest experiments that could provide further insights into
mechanisms of error discrimination and, more broadly, aid in our
understanding of recently discovered polymerases that exhibit
extraordinarily low fidelity .
binds to an incorrect nucleotide. Our
results suggest experiments that could provide further insights into
mechanisms of error discrimination and, more broadly, aid in our
understanding of recently discovered polymerases that exhibit
extraordinarily low fidelity .

 Click to go back to the publication list
Click to go back to the publication list
