Polymerase  Local Deformations Revealed by Dynamics Simulations of
DNA Polymerase
Local Deformations Revealed by Dynamics Simulations of
DNA Polymerase  with DNA Mismatches at the
Primer Terminus
with DNA Mismatches at the
Primer Terminus
Nanosecond dynamics simulations for DNA
polymerase  /DNA complexes with three mismatched
base pairs, either GG, CA, or CC (primer/template)
at the DNA polymerase active site,
are performed to investigate the mechanism of
polymerase opening and how the mispairs may affect
the DNA extension step; these trajectories
are compared to the behavior of a DNA polymerase
/DNA complexes with three mismatched
base pairs, either GG, CA, or CC (primer/template)
at the DNA polymerase active site,
are performed to investigate the mechanism of
polymerase opening and how the mispairs may affect
the DNA extension step; these trajectories
are compared to the behavior of a DNA polymerase  /DNA
complex with the correct base pair GC, and
assessed with the aid of targeted molecular
dynamics simulations of all systems from the
closed to the open enzyme state.
DNA polymerase conformational changes (subdomain
closing and opening) have been suggested to play
a critical role in DNA synthesis fidelity
since these changes are associated with the
formation of the substrate binding pocket for
the nascent base pair. Here we
observe different large C-terminal subdomain
(thumb) opening motions
in the simulations of DNA polymerase
/DNA
complex with the correct base pair GC, and
assessed with the aid of targeted molecular
dynamics simulations of all systems from the
closed to the open enzyme state.
DNA polymerase conformational changes (subdomain
closing and opening) have been suggested to play
a critical role in DNA synthesis fidelity
since these changes are associated with the
formation of the substrate binding pocket for
the nascent base pair. Here we
observe different large C-terminal subdomain
(thumb) opening motions
in the simulations of DNA polymerase 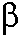 with GC versus GG base pairs. Whereas the
conformation of DNA polymerase
with GC versus GG base pairs. Whereas the
conformation of DNA polymerase 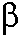 in the former approaches
the observed open state in the crystal structures,
the enzyme in the latter
does not. Interestingly, rotation of Arg258 toward
Asp192 which coordinates both active site metals
in the closed `active' complex, occurs rapidly
in the GG simulation. We have previously
suggested that this rotation is a key slow step
in the closed to open transition.
Targeted molecular dynamics simulations
also point to a unique pathway for Arg258 rotation
in the GG mispair complex.
Further structural analysis in the active site
reveals distorted geometries of all these mispair
complexes examined. The
hierarchy of these distortions (GG > CC > CA)
parallels the experimentally deduced inability
of DNA polymerase
in the former approaches
the observed open state in the crystal structures,
the enzyme in the latter
does not. Interestingly, rotation of Arg258 toward
Asp192 which coordinates both active site metals
in the closed `active' complex, occurs rapidly
in the GG simulation. We have previously
suggested that this rotation is a key slow step
in the closed to open transition.
Targeted molecular dynamics simulations
also point to a unique pathway for Arg258 rotation
in the GG mispair complex.
Further structural analysis in the active site
reveals distorted geometries of all these mispair
complexes examined. The
hierarchy of these distortions (GG > CC > CA)
parallels the experimentally deduced inability
of DNA polymerase 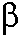 to extend these mispairs.
These studies on the DNA polymerase
to extend these mispairs.
These studies on the DNA polymerase 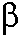 opening provide structural and dynamic insights,
helping to explain experimental results regarding
DNA extension following misincorporation.
opening provide structural and dynamic insights,
helping to explain experimental results regarding
DNA extension following misincorporation.

 Click to go back to the publication list
Click to go back to the publication list
