Madbend: a program for calculating the curvature of nucleic acid
Madbend is an acronym for the Macroscopic Analysis of DNA Bending. The program was authored by Daniel Strahs, a research specialist in the laboratory of Prof. Tamar Schlick. The first use and an example of Madbend's capabilities is provided in the companion paper "Analysis of A-tract bending: Insights into experimental structues by molecular dynamics simulations" (Strahs and Schlick, 2000).
Specifically, from a given list of base pair step roll, tilt and twist values, and the specification of a reference plane, the program produces the global tilt and global roll and the bend magnitude. These quantities thus describe the direction and magnitude of a bend angle and can be used to analyze DNA and DNA/protein complex geometries.
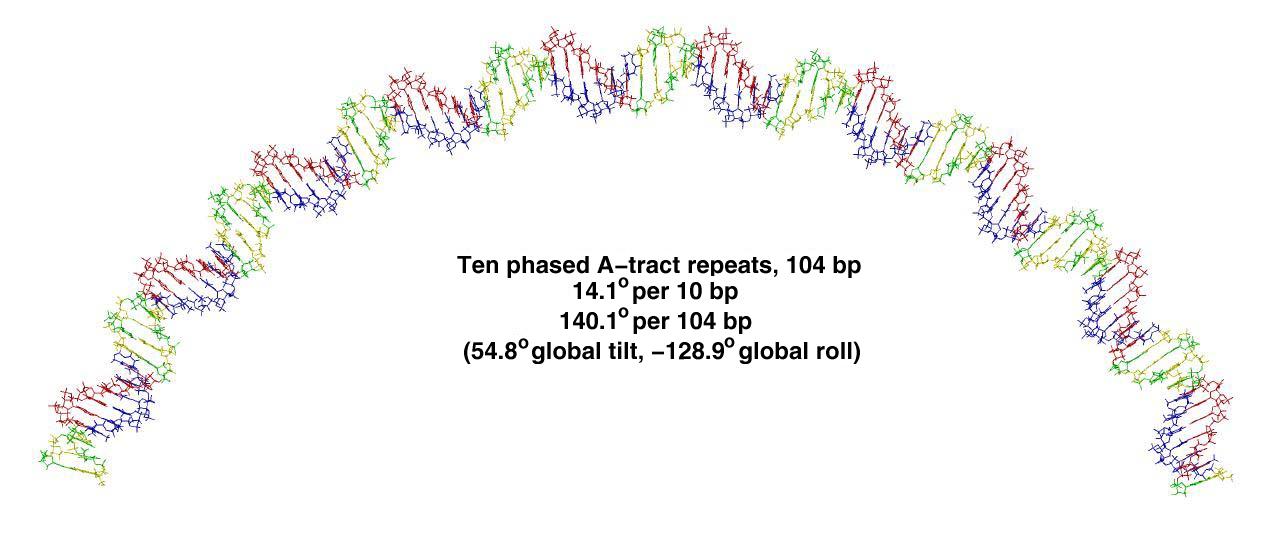
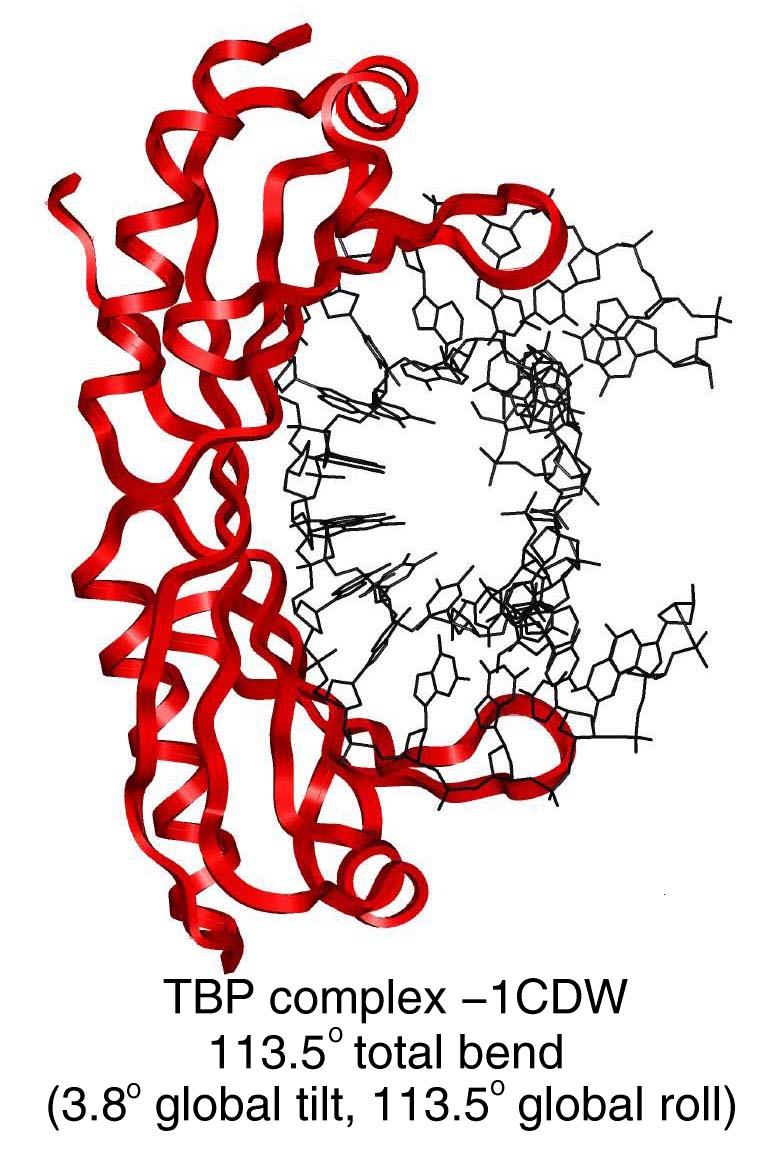
Madbend has several features including:
- Command line input, suitable for shell scripts
- Use of Curves output files as parameter input
- Exclusion syntax to assist in decomposing the overall bend from individual contributions and to eliminate end base pairs and artifacts
- Use of either output files or standard output
- Choice of verbose output
The program is a standalone utility. However, it requires the base pair step rotation parameters tilt, roll and twist as input parameters to calculate the bending of the oligonucleotide. Thus, Madbend is best paired with one of the many nucleic acid structure analysis programs (such as Curves, FREEHELIX, and RNA that are available.
License: By downloading this software, Licensee accepts a restricted, non-exclusive, non-transferable license to use the software for academic and research purposes only, e.g. not for commercial use without a fee. Should licensee wish to make commercial use of the software, Licensee will need to contact Dr. Tamar Schlick (schlick@nyu.edu) to negotiate an appropriate license.