|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Use the "FULL" & "ABS" icons to obtain the electronic versions of either the full length paper or it's abstract. A form at the bottom of the page allows you to request that a hard copy be mailed to you. If you forget what an icon means, click on any icon other than the "FULL" or "ABS" icons to see this table again. Using the "Back" button on your browser puts you back in the list.
***The manuscripts available on our site are provided for your personal use only and may not be retransmitted or redistributed without written permissions from the paper's publisher and author. You may not upload any of this site's material to any public server, on-line service, network, or bulletin board without prior written permission from the publisher and author. You may not make copies for any commercial purpose. Reproduction or storage of materials retrieved from this web site is subject to the U.S. Copyright Act of 1976, Title 17 U.S.C.***
-




Stephanie Portillo-Ledesma and Tamar Schlick, "Regulation of chromatin architecture by protein binding: insights from molecular modeling", Biophysical Reviews, https://doi.org/10.1007/s12551-024-01195-5, 2024. 



-




Giulio Quarta and Tamar Schlick, "Riboswitch Distribution in the Human Gut Microbiome Reveals Common Metabolite Pathways", J. Phys. Chem. B, https://doi.org/10.1021/acs.jpcb.4c00267, 2024. 


-




Tamar Schlick and Shuting Yan, "Modeling and Simulating RNA: Combining Structural, Dynamic, and Evolutionary Perspectives for Coronavirus Applications", Comprehensive Computational Chemistry, Yanez, Manuel and Boyd, Russell J. (eds.), http://dx.doi.org/10.1016/B978-0-12-821978-2.00118-5, 3:886-984, 2024. 



-





Zilong Li, and Tamar Schlick, "Hi-BDiSCO: folding 3D mesoscale genome structures from Hi-C data using brownian dynamics", Nucleic Acids Research, https://doi.org/10.1093/nar/gkad1121, 2023. 




-





Albert Mao, Carrie Chen, Stephanie Portillo-Ledesma, and Tamar Schlick, "Effect of Single-Residue Mutations on CTCF Binding to DNA: Insights from Molecular Dynamics Simulations", Int. J. Mol. Sci., https://doi.org/10.3390/ijms24076395, 24(7): 6395, 2023. 




-




Stephanie Portillo-Ledesma, Zilong Li, and Tamar Schlick, "Genome modeling: From chromatin fibers to genes", Curr. Opin. Struct. Biol., https://doi.org/10.1016/j.sbi.2022.102506, 78:102506, 2023. 




-




Shuting Yan, Qiyao Zhu, Jenna Hohl, Alex Dong, and Tamar Schlick, "Evolution of coronavirus frameshifting elements: Competingstem networks explain conservation and variability", Proc. Natl. Acad. Sci., https://doi.org/10.1073/pnas.2221324120, 120(20): e2221324120, 2023. 




-




Zilong Li, Stephanie Portillo-Ledesma, and Tamar Schlick, "Brownian Dynamics Simulations of Mesoscale Chromatin Fibers", Biophys. J., https://doi.org/10.1016/j.bpj.2022.09.013, 122:1-14, 2023. 




-




Tamar Schlick, "Innovations in biophysics: A sampling of ideas celebrating Ned Seeman's legacy", Biophys. J., https://doi.org/10.1016/j.bpj.2022.11.030, 121:E01-E02, 2022. 

-




Lewis Rolband, Damian Beasock, Yang Wang, Yao-Gen Shu, Jonathan D. Dinman, Tamar Schlick, Yaoqi Zhou, Jeffrey S. Kieft, Shi-Jie Chen, Giovanni Bussi, Abdelghani Oukhaled, Xingfa Gao, Petr Šulc, Daniel Binzel, Abhjeet S. Bhullar, Chenxi Liang, Peixuan Guo, Kirill A. Afonin, "Biomotors, viral assembly, and RNA nanobiotechnology: Current achievements and future directions", Computational and Structural Biotechnology Journal, https://doi.org/10.1016/j.csbj.2022.11.007, 20:6120-6137, 2022. 


-




Stephanie Portillo-Ledesma, Meghna Wagley and Tamar Schlick, "Chromatin transitions triggered by LH density as epigenetic regulators of the genome", Nucleic Acids Research, https://doi.org/10.1093/nar/gkac757, 50(18):10328-10342, 2022. 




-




Qiyao Zhu, Louis Petingi, and Tamar Schlick, "RNA-As-Graphs Motif Atlas—Dual Graph Library of RNA Modules and Viral Frameshifting-Element Applications", Int. J. Mol. Sci., https://doi.org/10.3390/ijms23169249, 23:9249, 2022. 



-





Shuting Yan, Qiyao Zhu, Swati Jain, and Tamar Schlick, "Length-dependent motions of SARS-CoV-2 frameshifting RNA pseudoknot and alternative conformations suggest avenues for frameshifting suppression", Nat Commun., https://doi.org/10.1038/s41467-022-31353-w, 13(1):4284, Jul 25, 2022. 




-




Sarah G Swygert, Dejun Lin, Stephanie Portillo-Ledesma, Po-Yen Lin, Dakota R Hunt, Cheng-Fu Kao, Tamar Schlick, William S Noble, and Toshio Tsukiyama, "Local chromatin fiber folding represses transcription and loop extrusion in quiescent cells", Elife, https://doi.org/10.7554/eLife.72062, November 4, 2021. 



-




Tamar Schlick, "From butterflies to bits: A sweeping vision for the code of life", Biophysical Reports, https://doi.org/10.1016/j.bpr.2021.100010, September 8, 2021 


-




Tamar Schlick, "Isabella L. Karle: A Crystallography Pioneer", DNA AND CELL BIOLOGY, DOI: 10.1089/dna.2021.0372, Volume 40, Number 7, 2021. 


-




Tamar Schlick and Jacob Fish, "MGO on the go: Multiscale genome symposium - annual biophysical society meeting 2021", Biophysical Reviews, 13:309–310 (2021) 


-




Tamar Schlick, Stephanie Portillo-Ledesma, Mischa Blaszczyk, Luke Dalessandro, Somnath Ghosh, Klaus Hackl, Cale Harnish, Shravan Kotha, Daniel Livescu, Arif Masud, Karel Matouš, Arturo Moyeda, Caglar Oskay, and Jacob Fish, "A Multiscale Vision - Illustrative Applications From Biology to Engineering", International Journal for Multiscale Computational Engineering, 19(2):39–73 (2021). 


-




Tamar Schlick, Stephanie Portillo-Ledesma, Christopher G Myers, Lauren Beljak, Justin Chen, Sami Dakhel, Daniel Darling, Sayak Ghosh, Joseph Hall, Mikaeel Jan, Emily Liang, Sera Saju, Mackenzie Vohr, Chris Wu, Yifan Xu, Eva Xue, "Biomolecular Modeling and Simulation: A Prospering Multidisciplinary Field", Annu Rev Biophys, doi: 10.1146/annurev-biophys-091720-102019, 50:267-301 (2021). 


-




Tamar Schlick and Stephanie Portillo-Ledesma, "Biomolecular modeling thrives in the age of technology", Nature Computational Science, https://doi.org/10.1038/s43588-021-00060-9 (2021). 


-




Pablo Aurelio Gómez-García, Stephanie Portillo-Ledesma, Maria Victoria Neguembor, Martina Pesaresi, Walaa Oweis, Talia Rohrlich, Stefan Wieser, Eran Meshorer, Tamar Schlick, Maria Pia Cosma, Melike Lakadamyali, "Mesoscale Modeling and Single-Nucleosome Tracking Reveal Remodeling of Clutch Folding and Dynamics in Stem Cell Differentiation", Cell Rep, doi: 10.1016/j.celrep.2020.108614, 34(2):108614 (2021). 



-




Tamar Schlick, Eric J Sundberg, Susan J Schroeder, M Madan Babu, "Biophysicists' outstanding response to Covid-19", Biophys J., doi: 10.1016/j.bpj.2021.02.020, 120(6):E1-E2 (2021). 

-




Qiyao Zhu and Tamar Schlick, "A Fiedler Vector Scoring Approach for Novel RNA Motif Selection", Journal of Physical Chemistry B, doi: https://doi.org/10.1021/acs.jpcb.0c10685, 125(4):1144–1155 (2021). 


-




Yusufova N, Kloetgen A, Teater M, Osunsade A, Camarillo JM, Chin CR, Doane AS, Venters BJ, Portillo-Ledesma S, Conway J, Phillip JM, Elemento O, Scott DW, Béguelin W, Licht JD, Kelleher NL, Staudt LM, Skoultchi AI, Keogh MC, Apostolou E, Mason CE, Imielinski M, Schlick T, David Y, Tsirigos A, Allis CD, Soshnev AA, Cesarman E, Melnick AM, "Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture", Nature, 589(7841):299-305 (2021). 



-




S. Portillo-Ledesma, LH. Tsao, M. Wagley, M. Lakadamyali, MP Cosma, and Tamar Schlick, " Nucleosome Clutches are Regulated by Chromatin Internal Parameters", J Mol Biol., 10.1016/j.jmb.2020.11.001 (2020). 



-




Tamar Schlick, Qiyao Zhu, Swati Jain, and Shutting Yan, "Structure-Altering Mutations of the SARS-CoV-2 Frameshifting RNA Element", Biophys J., doi: 10.1016/j.bpj.2020.10.012, 120(6):1040-1053 (2021). 



-




Evelyne Bischof, Jantine A.C. Broek, Charles R. Cantor, Ashley J. Duits, Alfredo Ferro, Hillary W. Gao, Zilong Li, Stella Luna de Maria, Naomi I. Maria, Bud Mishra, Kimberly I. Mishra, Lex van der Ploeg, Larry Rudolph, Tamar Schlick, & RxCOVEA Framework, "ANERGY TO SYNERGY−THE ENERGY FUELING THE RXCOVEA FRAMEWORK", International Journal for Multiscale Computational Engineering, 18(3): 329-333 (2020). 

-




Tamar Schlick, "Multiscale Genome Organization: Dazzling Subject and Inventive Methods", Biophysical Journal, doi: 10.1016/j.bpj.2020.04.007 (2020). 

-




Tamar Schlick, "Eight Suggestions for Future Leaders of Science and Technology", The Biophysicist, 1(1):1-5 (2020). 

-




Tamar Schlick, "My blue whale: Seeking order in a chaotic world. An autobiographical reflection", Proceedings of the Seventh International Conference on Algorithms and Computational Biology (AlCoB 2020), Springer Lecture Notes in Bioinformatics (LNBIS)/Lecture Notes in Computer Science (LNCS) 12099: pages ix–xvi, C. Martin-Vide, M. A. Vega-Rodrigez, and T. Wheeler (Eds.), (2020). 

-





Akshay Sridhar, Stephen E. Farr, Guillem Portella, Tamar Schlick, Modesto Orozco, and Rosana Collepardo-Guevara, "Emergence of chromatin hierarchical loops from protein disorder and nucleosome asymmetry", PNAS, doi: 10.1073/pnas.1910044117 (2020). 


-





Swati Jain, Qiyao Zhu, Amiel S.P. Paz, and Tamar Schlick, "Identification of novel RNA design candidates by clustering the extended RNA-As-Graphs library", Biochim Biophys Acta Gen Subj, 1864(6): 129534, doi: 10.1016/j.bbagen.2020.129534 (2020). 


-





Swati Jain, Yunwen Tao, and Tamar Schlick, "Inverse folding with RNA-As-Graphs produces a large pool of candidate sequences with target topologies", J. Struct. Biol, 209(6): 107438, doi: 10.1016/j.jsb.2019.107438 (2020). 


-





Stephanie Portillo, and Tamar Schlick, "Bridging chromatin structure and function over a range of experimental spatial and temporal scales by molecular modeling", WIREs Computational Molecular Science, 10(2): e1434 (2019). 


-




Louis Petingi, and Tamar Schlick, "Graph-Theoretic Partitioning of RNAs and Classification of Pseudoknots", Algorithms for Computational Biology, 68-79 (2019). 


-





Christopher G. Myers, Donald E. Olins, Ada L. Olins, and Tamar Schlick, "Mesoscale Modeling of Nucleosome-Binding Antibody PL2-6: Mono- versus Bivalent Chromatin Complexes", Biophys J, (118): 1-11, doi: 10.1016/j.bpj.2019.08.019 (2019). 



-





Grace Meng, Marva Tariq, Swati Jain, Shereef Elmetwaly, and Tamar Schlick, "RAG-Web: RNA Structure Prediction/Design using RNA-As-Graphs", Bioinformatics, doi: 10.1093/bioinformatics/btz611 (2019). 


-




Jaewoon Jung, Wataru Nishima, Marcus Daniels, Gavin Bascom, Chigusa Kobayashi, Adetokunbo Adedoyin, Michael Wall, Anna Lappala, Dominic Phillips, William Fischer, Chang‐Shung Tung, Tamar Schlick, Yuji Sugita, and Karissa Y. Sanbonmatsu, "Scaling molecular dynamics beyond 100,000 processor cores for large-scale biophysical simulations", Journal of Computational Chemistry, 40(21): 1919-1930 (2019). 

-





Ognjen Perisic, Stephanie Portillo-Ledesma, and Tamar Schlick, "Sensitive effect of linker histone binding mode and subtype on chromatin condensation", Nucleic Acids Research, doi: 10.1093/nar/gkz234 (2019). 



-





Swati Jain, Sera Saju, Louis Petingi, and Tamar Schlick, "An Extended Dual Graph Library and Partitioning Algorithm Applicable to Pseudoknotted RNA Structures", Methods, doi: 10.1016/j.ymeth.2019.03.022 (2019). 


-





Gavin Bascom, Christian Myers, and Tamar Schlick, "Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub", PNAS, 116(11): 4955-4962 (2019). 



-





Swati Jain, Cigdem S. Bayrak, Louis Petingi, and Tamar Schlick, "Dual Graph Partitioning Highlights a Small Group of Pseudoknot-Containing RNA Submotifs", Genes, 9(8): 371 (2018). 


-





Swati Jain, Alain Laederach, Silvia B V. Ramos, and Tamar Schlick, "A pipeline for computational design of novel RNA-like topologies", Nucleic Acids Research, 46(14): 7040-7051, doi.org/10.1093/nar/gky524 (2018). 


-




Gavin Bascom, and Tamar Schlick, "Chromatin Fiber Folding Directed by Cooperative Histone Tail Acetylation and Linker Histone Binding", Biophysical Journal, 114(10): 2376-2385 (2018). 



-




Tamar Schlick, "Adventures with RNA graphs", Methods, 143: 16-33, doi: 10.1016/j.ymeth.2018.03.009 (2018). 


-



Gavin Bascom, and Tamar Schlick, "Mesoscale Modeling of Chromatin Fibers", Nuclear Architecture and Dynamics, 123-147 (2018). 


-




Swati Jain, and Tamar Schlick, "F-RAG: Generating Atomic Coordinates from RNA Graphs by Fragment Assembly", J. Mol. Biol., 429(23): 3587-3605 (2017). 


-




Suhas S.P. Rao, Su-Chen Huang, Brian Glenn St Hilaire, Jesse M. Engreitz, Elizabeth M. Perez, Kyong-Rim Kieffer-Kwon, Adrian L. Sanborn, Sarah E. Johnstone, Gavin D. Bascom, Ivan D. Bochkov, Xingfan Huang, Muhammad S. Shamim, Jaeweon Shin, Douglass Turner, Ziyi Ye, Arina D. Omer, James T. Robinson, Tamar Schlick, Bradley E. Bernstein, Rafael Casellas, Eric S. Lander, and Erez Lieberman Aiden, "Cohesin Loss Eliminates All Loop Domains", Cell, 171, 305-320 (2017). 



-





Ognjen Perisic, and Tamar Schlick, "Dependence of the Linker Histone and Chromatin Condensation on the Nucleosome Environment", The Journal of Physical Chemistry-B, 121(33): 7823-7832 (2017). 

-




Tamar Schlick, and Anna Marie Pyle, "RNA Structural Variability and Functional Versatility Challenge RNA Structural Modeling and Design", Biophysical Journal, 113(2): E1-E2 (2017). 

-





Gavin Bascom, Taejin Kim, and Tamar Schlick, "Kilobase Pair Chromatin Fiber Contacts Promoted by Living-System-Like DNA Linker Length Distributions and Nucleosome Depletion", The Journal of Physical Chemistry-B, 121(15): 3882-3894 (2017). 



-




Tamar Schlick, and Anna Marie Pyle, "Opportunities and Challenges in RNA Structural Modeling and Design", Biophysical Journal, 113(2): 225-234 (2017). 

-





Cigdem Sevim Bayrak, Namhee Kim, and Tamar Schlick, "Using sequence signatures and kink-turn motifs in knowledge-based statistical potentials for RNA structure prediction", Nucleic Acids Research, 45(9): 5414-5422 (2017). 

-




Gavin Bascom, and Tamar Schlick, "Linking Chromatin Fibers to Gene Folding by Hierarchical Looping", Biophysical Journal, 112(3): 434-445 (2017). 


-




Tamar Schlick and Leslie Loew, "Unraveling Genome Biophysics", Biophysical Journal, 112(3): E1-E2 (2017). 

-




Ognjen Perisic, and Tamar Schlick, "Computational strategies to address chromatin structure problems", Phys Biol., 13(3): 035006 (2016). 

-





Antoni Luque, Gungor Ozer, and Tamar Schlick, "Correlation among DNA Linker Length, Linker Histone Concentration, and Histone Tails in Chromatin", Biophysical Journal, 110(11): 2309-2319 (2016). 



-





Gavin Bascom, Karissa Y Sanbonmatsu, and Tamar Schlick, "Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements", The Journal of Physical Chemistry-B, 120(33): 8642-53 (2016). 


-





Taejin Kim, Bret D. Freudenthal, William A. Beard, Samuel H. Wilson, and Tamar Schlick, "Insertion of oxidized nucleotide triggers rapid DNA polymerase opening", Nucleic Acids Research, 44(9): 4409-24, doi: 10.1093/nar/gkw174 (2016). 


-




Anna Marie Pyle, and Tamar Schlick, "Challenges in RNA Structural Modeling and Design", J. Mol. Biol., 428 (5):733-735, doi: 10.1016/j.jmb.2016.02.012 (2016). 
-




Sergei Grigoryev, Gavin Bascom, Jenna Buckwalter, Michael Schubert, Christopher Wookcock, and Tamar Schlick, "Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes", PNAS, 113(5):1238-43, doi: 10.1073/pnas.1518280113 (2016). 



-




Lei Hua, Yang Song, Namhee Kim, Christian Laing, Jason Wang, and Tamar Schlick, "CHSalign: A Web Server That Builds upon Junction-Explorer and RNAJAG for Pairwise Alignment of RNA Secondary Structures with Coaxial Helical Stacking", PLoS ONE, 11(1):e0147097, DOI: 10.1371/journal.pone.0147097 (2016). 


-




Louis Petingi, and Tamar Schlick, "Partitioning RNAs into pseudonotted and pseudoknot-free regions modeled as Dual Graphs", q-bio.QM,arXiv:1601.04259 (2016). 
-





Naoto Baba, Shereef Elmetwaly, Namhee Kim, and Tamar Schlick, "Predicting Large RNA-Like Topologies by a Knowledge-Based Clustering Approach", J. Mol. Biol., 428(5 Pt A): 811-21, doi: 10.1016/j.jmb.2015.10.009 (2015). 

-




Mai Zahran, Cigdem Sevim Bayrak, Shereef Elmetwaly, and Tamar Schlick, "RAG-3D: a search tool for RNA 3D substructures", Nucleic Acids Research, 43(19): 9474-88, doi: 10.1093/nar/gkv823 (2015). 


-





Rosana Collepardo-Guevara, Guillem Portella, Michele Vendruscolo, Daan Frenkel, Tamar Schlick, and Modesto Orozco, "Chromatin Unfolding by Epigenetic Modifications Explained by Dramatic Impairment of Internucleosome Interactions: A Multiscale Computational Study", Journal of the American Chemical Society, 137:(32), 10205-10215 (2015). 


-




Gungor Ozer, Antoni Luque, and Tamar Schlick, "The chromatin fiber: multiscale problems and approaches", Curr. Opin. Struct. Biol., 31:, 124-139 (2015). 



-




Namhee Kim, Mai Zahran, and Tamar Schlick, "Computational Prediction of Riboswitch Tertiary Structures Including Pseudoknots by RAGTOP: A Hierarchical Graph Sampling Approach", Methods Enzymol., 553, 115-35, DOI: 10.1016/bs.mie.2014.10.054 (2015). 

-




Gungor Ozer, Rosana Collepardo-Guevara, and Tamar Schlick, "Forced unraveling of chromatin fibers with nonuniform linker DNA lengths", J. Phys.: Condens. Matter, 27(6): 064113, DOI: 10.1088/0953-8984/27/6/064113 (2015). 



-




Bret D. Freudenthal, William A. Beard, Lalith Perera, David D. Shock, Taejin Kim, Tamar Schlick, and Samuel H. Wilson, "Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide", Nature, 517(7536): 635–639, DOI:10.1038/nature13886 (2014). 




-




Namhee Kim, Zhe Zheng, Shereef Elmetwaly and Tamar Schlick, "RNA Graph Partitioning for the Discovery of RNA Modularity: A Novel Application of Graph Partition Algorithm to Biology", PLoS ONE, 9(9): e106074, DOI: 10.1371/journal.pone.0106074 (2014). 


-





Antoni Luque, Rosana Collepardo-Guevara, Sergei Grigoryev and Tamar Schlick, "Dynamic condensation of linker histone C-terminal domain regulates chromatin structure", Nucleic Acids Res., 42(12): 7553-7560, DOI: 10.1093/nar/gku491 (2014). 



-





Rosana Collepardo-Guevara, and Tamar Schlick, "Chromatin fiber polymorphism triggered by variations of DNA linker lengths", PNAS, DOI: 10.1073/pnas.1315872111 (2014). 



-





Namhee Kim, Christian Laing, Shereef Elmetwaly, Segun Jung, Jeremy Curuksu, and Tamar Schlick, "Graph-based sampling for approximating global helical topologies of RNA", PNAS, DOI: 10.1073/pnas.1318893111 (2014). 


-



Yunlang Li, Bret D. Freudenthal, William A. Beard, Samuel H. Wilson, and Tamar Schlick, "Optimal and Variant Metal-Ion Routes in DNA Polymerase β's Conformational Pathways", J. Am. Chem. Soc.,DOI: 10.1021/ja412701f (2014). 


-




Segun Jung and Tamar Schlick, "Interconversion between Parallel and Antiparallel Conformations of a 4H RNA junction in Domain 3 of Foot-and-Mouth Disease Virus IRES Captured by Dynamics Simulations", Biophys. J., 106(2): 447-458, (2014). 


-





Benedetta Sampoli Benítez, Zachary R. Barbati, Karunesh Arora, Jasmina Bogdanovic, and Tamar Schlick, "How DNA Polymerase X Preferentially Accommodates Incoming dATP Opposite 8-Oxoguanine on the Template", Biophys. J., 105(11): 2559-2568, (2013). 


-




Tamar Schlick, "The 2013 Nobel Prize in Chemistry Celebrates Computations in Chemistry and Biology", SIAM News, 46(10), December (2013). -





Christian Laing, Segun Jung, Namhee Kim, Shereef Elmetwaly, Mai Zahran, and Tamar Schlick, "Predicting Helical Topologies in RNA Junctions as Tree Graphs", PLoS ONE, 8(8): e71947, DOI:10.1371/journal.pone.0071947 (2013). 


-





Yunlang Li, and Tamar Schlick, "Gate-keeper Residues and Active-Site Rearrangements in DNA Polymerase μ Help Discriminate Non-cognate Nucleotides", PLoS Comput. Biol., 9(5), DOI:10.1371/journal.pcbi.1003074 (2013). 


-




Rosana Collepardo-Guevara, and Tamar Schlick, "Insights into chromatin fibre structure by in vitro and in silico single-molecule stretching experiments", Biochem. Soc. Trans., 41(2): 494-500 (2013). 


-




Segun Jung, and Tamar Schlick, "Candidate RNA structures for domain 3 of the foot-and-mouth-disease virus internal ribosome entry site", Nucleic Acids Res., 41(3): 1483-95 (2013). 


-




Namhee Kim, Louis Petingi, and Tamar Schlick, "Network Theory Tools for RNA Modeling", WSEAS Transactions on Math, 12(9): 941-955 (2013). 


-




Namhee Kim, Niccole Fuhr and Tamar Schlick, "Graph Applications to RNA Structure and Function", Chapter 3, pp. 23–51, In Biophysics of RNA Folding, Ed. R. Russel, Biophysics for the Life Sciences 3, Springer Verlag (2013). 


-




Rosana Collepardo-Guevara, and Tamar Schlick, "Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes", Nucleic Acids Res., 40(18): 8803-8817 (2012). 


-




Tamar Schlick, Karunesh Arora, William A. Beard, and Samuel H. Wilson, "Perspective: pre-chemistry conformational changes in DNA polymerase mechanisms", Theor Chem Acc, 131(12): 1287 (2012). 


-




Namhee Kim and Tamar Schlick, "A New Toolkit for Modeling RNA from a Pseudo-Torsional Space", J. Mol. Biol., doi:10.1016/j.jmb.2012.05.027 (2012). 


-





Yunlang Li, Chelsea L. Gridley, Joachim Jaeger, Joann B. Sweasy, and Tamar Schlick, "Unfavorable electrostatic and steric interactions in DNA polymerase β E295K mutant interfere with the enzyme’s pathway", J. Am. Chem. Soc., 134(24): 9999-10010 (2012). 


-




Tamar Schlick, "Modelers to the Rescue", In Innovations in Biomolecular Modeling and Simulations, Volume 1, T. Schlick, ed., Royal Society of Chemistry, London (2012). -




Meredith Foley, Karunesh Arora and Tamar Schlick, "Intrinsic Motions of DNA Polymerases Underlie Their Remarkable Specificity and Selectivity and Suggest a Hybrid Substrate Binding Mechanism", In Innovations in Biomolecular Modeling and Simulations, Volume 2, T. Schlick, ed., Royal Society of Chemistry, London (2012). 


-



Innovations in Biomolecular Modeling and Simulations, Volume 2, T. Schlick ed., The Royal Society of Chemistry, ISBN: 978-1-84973-505-6, DOI:10.1039/9781849735056 (2012). -



Innovations in Biomolecular Modeling and Simulations, Volume 1, T. Schlick ed., The Royal Society of Chemistry, ISBN: 978-1-84973-504-9, DOI:10.1039/9781849735049 (2012). -




Tamar Schlick, Jeff Hayes, and Sergei Grigoryev, "Toward Convergence of Experimental Studies and Theoretical Modeling of the Chromatin Fiber", JBC, 287: 5183-5191 (2012). 

-




Giulio Quarta, Ken Sin and Tamar Schlick, "Dynamic Energy Landscapes of Riboswitches Help Interpret Conformational Rearrangements and Function", PLoS Comput Biol, 8(2): e1002368. doi:10.1371/journal.pcbi.1002368 (2012). 

-





Rosana Collepardo-Guevara and Tamar Schlick, "The Effect of Linker Histone's Nucleosome Binding Affinity on Chromatin Unfolding Mechanisms", Biophys. J., 111(7):1670-1680 (2011). 


-




Christian Laing, Dongrong Wen, Jason T. L. Wang and Tamar Schlick, "Predicting coaxial helical stacking in RNA junctions", Nucleic Acids Research, 40(2): 487-498, doi:10.1093/nar/gkr629 (2011). 

-




Tamar Schlick and Rosana Collepardo-Guevara, "Biomolecular Modeling and Simulation: The Productive Trajectory of a Field", SIAM News, 44: 6 (2011). -




Joseph Izzo, Namhee Kim, Shereef Elmetwaly and Tamar Schlick, "RAG: an update to RNA-As-Graphs resource", BMC Bioinformatics, 12: 219 (2011). 

-




Christian Laing and Tamar Schlick, "Computational approaches to RNA structure prediction, analysis, and design", Curr. Opin. Struct. Biol., 21: 1-13 (2011). 

-




Tamar Schlick, "DNA Polymerases: Structure, Function, and Modeling", in Molecular Machines, , B. Roux, Editor, World Scientific, (2011). 



-





Yunlang Li and Tamar Schlick, "Modeling DNA Polymerase μ Motions: Subtle Transitions before Chemistry", Biophys. J., 99(10): 3463-3472 (2010). 



-





Hin Hark Gan and Tamar Schlick, "Chromatin Ionic Atmosphere Analyzed by a Mesoscale Electrostatic Approach", Biophys. J., 99(8): 2587-2596 (2010). 

-




Tamar Schlick, Rosana Collepardo-Guevara, Leif A. Halvorsen, Segun Jung and Xia Xiao, "Biomolecular Modeling and Simulation: A Field Coming of Age" , Q. Rev. of Biophys., 44 (2): 191-228 (2011). 



-




Meredith Foley, Victoria Padow and T. Schlick, "Extraordinary Ability of DNA Pol λ to Stabilize Misaligned DNA" , J. Am. Chem. Soc., 132: 13403-13416 (2010). 



-





Ognjen Perisic, Rosana Collepardo-Guevara and T. Schlick, "Modeling Studies of Chromatin Fiber Structure as a Function of DNA Linker Length", J. Mol. Biol., 403: 777-802 (2010). 



-





N. Kim, J. Izzo, S. Elmetwaly, H.H. Gan and T. Schlick, "Computational Generation and Screening of RNA Motifs in Large Nucleotide Sequence Pools", Nucleic Acids Research, doi:10.1093/nar/gkq282 (2010). 


-




C. Laing, and T. Schlick, "Computational Approaches to RNA 3D Modeling", J. Phys.: Condens. Matter, 22: 283101 (2010). 


-





G. Quarta, N. Kim, J. A. Izzo, and T. Schlick, "Analysis of Riboswitch Structure and Function by an Energy Landscape Framework.", J. Mol. Biol., 393:993-1003 (2009). 



-





C. Laing and T. Schlick, "Analysis of Four-Way Junctions in RNA Structures.", J. Mol. Biol., 390:547-559 (2009). 



-




Tamar Schlick, "Mathematical and Biological Scientists Assess the State-of-the-Art in RNA Science at an IMA Workshop RNA in Biology, Bioengineering and Biotechnology", Int. J. Multi. Sci. Eng., 8 (4):369-378 (2010) 



-




Tamar Schlick and Ognjen Perisic, "Mesoscale simulations of two nucleosome-repeat length oligonucleosomes.", Phys. Chem. Chem. Phys., 11:10729-10737 (2009) 



-




S. Grigoryev, G. Arya, S. Correll, C. Woodcock, and T. Schlick, "Evidence for hetromorphic chromatin fibers from analysis of nucleosome interactions.", PNAS, 106(32):13317-13322 (2009) 



-




C. Laing, S. Jung, A. Iqbal, and T. Schlick, "Tertiary motifs revealed in analyses of higher-order RNA junctions.", J. Mol. Biol., 393(1):67-82 (2009) 



-




T. Schlick, "From Macroscopic to Mesoscopic Models of Chromatin Folding." Chapter 15, pp. 514--535, Multiscale Methods: Bridging the Scales in Science and Engineering, J. Fish, Editor, Oxford University Press (2009) 



-




T. Schlick, "Molecular dynamics-based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules." F1000 Biology Reports, 1-51 (2009) 



-





M.C. Foley and T. Schlick, "Relationship Between Conformational Changes in Pol λ Active Site Upon Binding Incorrect Nucleotides and Mismatch Incorporation Rates" J. Phys. Chem. B, 113:13035-13047 (2009) 



-




T. Schlick, "Monte Carlo, harmonic approximation, and coarse-graining approaches for enhanced sampling of biomolecular structure." F1000 Biology Reports, 1-48(2009) 



-




G. Arya and T. Schlick, "A Tale of Tails: How Histone Tails Mediate Chromatin Compaction in Different Salt and Linker Histone Environments" J. Phys. Chem. A, 16:4045-4059 (2009) 


-




B. A. Sampoli Benitez, K. Arora, L. Balistreri, and T. Schlick, "Mismatched base-pair simulations for ASFV Pol X/DNA complexes help interpret frequent G*G misincorporation", J. Mol. Biol., 384:1086-1097 (2008) 



-




P. Sridhar, H.H. Gan, and T. Schlick, "A computational screen for C/D box snoRNAs in the human genomic region associated with Prader-Willi and Angelman syndromes", J. Biomed. Sci., 15:697-705 (2008) 



-





K. Bebenek, M. Garcia-Diaz, M. C. Foley, L. C. Pedersen, T. Schlick, and T. A. Kunkel, "Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase λ", EMBO Rep., 9:459-464 (2008) 



-




Y. Wang and T. Schlick, "Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization", J. Am. Chem. Soc., 130:13240-13250 (2008) 




-




M. C. Foley and T. Schlick, "Simulations of DNA pol λ R517 mutants indicate 517's crucial role in ternary complex stability and suggest DNA slippage origin", J. Am. Chem. Soc., 130:3967-3977 (2008) 




-





Y. Xin, C. Laing, N.B. Leontis, and T. Schlick, "Annotation of tertiary interactions in RNA structures reveals variations and correlations.", RNA, 14:2465-2477 (2008) 

-




Y. Xin, G. Quarta, H.H. Gan, and T. Schlick, "Estimating the fraction of noncoding RNAs in mammalian transcriptomes.", Bioinformatics and Biology Insights, 2:77-95 (2008) 

-




N. Kim, J.S. Shin, S. Elmetwaly, H.H. Gan and T. Schlick, "RAGPOOLS: RNA-As-Graph-Pools A Web Server for Assisting the Design of Structured RNA Pools for In Vitro Selection", Bioinformatics, 23(21):2959-2960, doi:10.1093/bioinformatics/btm439 (2007) 

-




M.D. Bojin and T. Schlick, "A quantum mechanical investigation of possible mechanisms for the nucleotidyl transfer reaction catalyzed by DNA polymerase 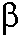 ", J. Phys. Chem. B, 111(38),11244-11252 (2007)
", J. Phys. Chem. B, 111(38),11244-11252 (2007) 



-




I.L. Alberts, Y. Wang, T. Schlick, "DNA polymerase 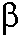 catalysis: are different mechanisms possible?" , J Am Chem Soc, 129(36):11100-10 (2007)
catalysis: are different mechanisms possible?" , J Am Chem Soc, 129(36):11100-10 (2007) 




-




N. Kim, H. H. Gan, and T. Schlick, "A Computational Proposal for Designing Structured RNA Pools for in Vitro Selection of RNAs", RNA, 13:478-92 (2007) 




-




Y. Wang, S. Reddy, W. Beard, S. Wilson, and T. Schlick, "Differing Conformational Pathways Before and After Chemistry for Insertion of dATP vs. dCTP Opposite 8-oxoG in DNA Polymerase Beta", Biophys. J.,92:3063-3070 (2007) 


-




Y. Wang and T. Schlick, "Distinct Energetics and Closing Pathwasys for DNA Polymerase Beta with 8-oxoG Template and Different Incoming Nucleotides" BMC Structural Biology, 7:7 (2007) 




-




G. Arya and T. Schlick, "Efficient Global Biopolymer Sampling with End-Transfer Configurational Bias Monte Carlo" J. Chem. Phys., 126:044107 (2007) 


-




R. Radhakrishnan and T. Schlick, "Correct and Incorrect Nucleotide Incorporation Pathways in DNA Polymerase Betas" Biochem. Biophys. Res. Comm., 350:521-529 (2006) 






-




M. Foley, K. Arora, and T. Schlick, "Sequential Side-chain Residue Motions Transform the Binary into the Ternary State of DNA Polymerase Lambda" Biophys. J., 91:3182-3195 (2006) 



-




G. Arya and T. Schlick, "Role of Histone Tails in Chromatin Folding Revealed by a New Mesoscopic Oligonucleosome Model", PNAS, 103:16236-16241 (2006) 


-





R. Radhakrishnan, K. Arora, Y. Wang, W. Beard, S. Wilson, and T. Schlick, "Regulation of DNA Repair Fidelity by Molecular Checkpoints: "Gates" in DNA Polymerase Beta's Substrate Selection", Biochem., 45:15142-15156 (2006) 



-




G. Arya, Q. Zhang, and T. Schlick, "Flexible Histone Tails in a New Mesopscopic Model of Oligonucleosomes", Biophys. J., 91:133-150 (2006) 


-




Q. Zhang and T. Schlick, "Stereochemistry and Position-Dependent Effects of Carcinogens on TATA/TBP Binding", Biophys. J., 90:1865-1877 (2006) 


-




T. Schlick, "RNA: The Cousin Left Behind Becomes a Star", in Computational Studies of DNA and RNA, pp.259-281, J. Sponer and F. Lankas, Editors, Springer Verlag, Dordrecht, The Netherlands (2006) -




B. Sampoli Benitez, K. Arora, and T. Schlick, "In Silico Studies of the African Swine Fever Virus DNA Polymerase X support an Induced-fit Mechanism", Biophys. J., 90:42-56 (2006) 



-




Y. Wang, K. Arora, and T. Schlick, "Subtle but Variable Conformational Rearrangements in the Replication Cycle of Sulfolobus solfactarius P2 DNA polymerase IV (Dpo4) May Accommodate Lesion Bypass", Prot. Sci., 15:135-151 (2006) 



-




K. Arora, W. A. Beard, S. H. Wilson, and T. Schlick, "Mismatch Induced Conformational Distortions in Polymerase 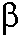 Support an Induced-Fit Mechanism for Fidelity", Biochem., 44:13328-13341 (2005)
Support an Induced-Fit Mechanism for Fidelity", Biochem., 44:13328-13341 (2005) 


-




U. Laserson, H. H. Gan, and T. Schlick, "Predicting Candidate Genomic Sequences that Correspond to Synthetic Functional RNA Motifs", Nuc. Acids Res., 33:6057-6069 (2005) 



-




U. Laserson, H. H. Gan, and T. Schlick, "Exploring the Connection Between Synthetic and Natural RNAs in Genomes Via a Novel Computational Approach", in Advances in Algorithms for Macromolecular Simulation, Proceedings of the Fourth International Workshop on Algorithms for Macromolecular Modelling, Leicester, UK, August 2004, C. Chipot, R. Elber, A. Laaksonen, B. Leimkuhler, A. Mark, T. Schlick, R.D. Skeel, C. Schuette, Editors, Lecture Notes in Computational Science and Engineering, Vol. 49, pp. 35-56, Springer-Verlag, Berlin (2005) 



-





J. Gevertz, H. H. Gan, and T. Schlick, " In Vitro RNA Random Pools are Not Structurally Diverse: A Computational Analysis", RNA, 11:853-863 (2005) 



-




R. Radhakrishnan and T. Schlick, "Fidelity Discrimination in DNA Polymerase 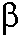 : Differing Closing Profiles for a Mismatched (G:A) Versus Matched (G:C) Base Pair", J. Amer. Chem. Soc., 127:13245-13252 (2005)
: Differing Closing Profiles for a Mismatched (G:A) Versus Matched (G:C) Base Pair", J. Amer. Chem. Soc., 127:13245-13252 (2005) 


-




J. Sun, Q. Zhang, and T. Schlick, "Electrostatic Mechanism of Nucleosomal Array Folding Revealed by Computer Simulation", PNAS, 102:8180-8185 (2005) 
-




K. Arora and T. Schlick, "Conformational Transition Pathway of Polymerase 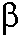 /DNA Complex Upon Binding Correct Incoming Substrate", J. Phys. Chem. B, 109:5358-5367 (2005)
/DNA Complex Upon Binding Correct Incoming Substrate", J. Phys. Chem. B, 109:5358-5367 (2005) 


-




T. Schlick, "The Critical Collaboration Between Art and Science: Applying an Experiment on a Bird in an Air Pump to the Ramifications of Genomics on Society", Leonardo, 38(4):323-329 (2005) -





S. Pasquali, H. H. Gan, and T. Schlick, "Modular RNA Architecture Revealed by Computational Analysis of Existing Pseudoknots and Ribosomal RNAs", Nuc. Acids Res., 33:1384-1398 (2005) 


-





N. Kim, N. Shiffeldrim, H. H. Gan, and T. Schlick, "Candidates for Novel RNA Topologies", J. Mol. Biol., 341:1129-1144 (2004) 


-




R. Radhakrishnan and T. Schlick, "Biomolecular Free Energy Profiles by a Shooting/Umbrella Sampling Protocol, `BOLAS' ", J. Chem. Phys., 121:2436-2444 (2004) 


-




U. Laserson, H. H. Gan, and T. Schlick, "Searching for 2D RNA Geometries in Bacterial Genomes", Proceedings of the Twentieth Annual ACM Symposium on Computational Geometry, June 9-11, New York, 373-377, ACM Press (2004) 



-




R. Radhakrishnan and T. Schlick, "Orchestration of Cooperative Events in DNA Synthesis and Repair Mechanism Unraveled by Transition Path Sampling of DNA Polymerase 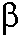 's Closing", Proc. Natl. Acad. Sci., 101:5970-5975 (2004)
's Closing", Proc. Natl. Acad. Sci., 101:5970-5975 (2004) 


-




L. Yang, K. Arora, W. A. Beard, S. H. Wilson, and T. Schlick, "Critical Role of Magnesium Ions in DNA Polymerase 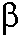 's Closing and Active Site Assembly", J. Am. Chem. Soc., 126:8441-8453 (2004)
's Closing and Active Site Assembly", J. Am. Chem. Soc., 126:8441-8453 (2004) 



-





K. Arora and T. Schlick, "In Silico Evidence for DNA Polymerase 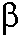 's Substrate-Induced Conformational Change", Biophys. J., 87:3088-3099 (2004)
's Substrate-Induced Conformational Change", Biophys. J., 87:3088-3099 (2004) 


-





K. Arora and T. Schlick, "Deoxyadenosine Sugar Puckering Pathway Simulated by the Stochastic Difference Equation Algorithm", Chem. Phys. Lett., 378:1-8 (2003) 


-





Q. Zhang, S. Broyde, and T. Schlick, "Deformations of Promoter DNA Bound to Carcinogens Help Interpret Effects on TATA-element Structure and Activity", Phil. Trans. Roy. Soc. Lond., Series A: Mathematical, Physical & Engineering Sciences, 362: 1479-1496 (2004) 



-




H. H. Gan, D. Fera, J. Zorn, N. Shiffeldrim, U. Laserson, N. Kim, and T. Schlick, "RAG: RNA-As-Graphs Database - Concepts, Analysis, and Features", Bioinformatics, 20: 1285-1291 (2004) 



-




D. Fera, N. Kim, N. Shiffeldrim, J. Zorn, U. Laserson, H. H. Gan, and T. Schlick, "RAG: RNA-As-Graphs Web Resource", BMC Bioinformatics, 5:88 (2004) 



-




J. Zorn, H. H. Gan, N. Shiffeldrim, and T. Schlick, "Structural Motifs in Ribosomal RNAs: Implications for RNA Design and Genomics", Biopolymers, 73: 340-347 (2004) 


-




Q. Zhang, D. A. Beard, and T. Schlick, "Constructing Irregular Surfaces to Enclose Macromolecular Complexes for Mesoscale Modeling Using the Discrete Surface Charge Optimization (DiSCO) Algorithm", J. Comp. Chem., 24:2063-2074 (2003) 

-



T. Schlick, "Engineering Teams Up with Computer-Simulation and Visualization Tools to Probe Biomolecular Mechanisms", Biophys. J., New and Notable, 85:1-4 (2003) 


-




L. Yang, W. A. Beard, S. H. Wilson, S. Broyde, and T. Schlick, "Highly Organized but Pliant Active-site of DNA Polymerase 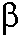 : Compensatory Mechanisms in Mutant Enzymes Revealed by Dynamics Simulations and Energy Analysis", Biophys. J., 86: 3392-3408 (2004)
: Compensatory Mechanisms in Mutant Enzymes Revealed by Dynamics Simulations and Energy Analysis", Biophys. J., 86: 3392-3408 (2004) 



-




D. A. Beard and T. Schlick, "Unbiased Rotational Moves for Rigid-Body Dynamics", Biophys. J., 85:2973-2976 (2003) -


H. H. Gan and T. Schlick, "Methods for Macromolecular Modeling (M3): Assessment of Progress and Future Perspectives", in Computational Methods for Macromolecules: Challenges and Applications - Proceedings of the 3rd International Workshop on Algorithms for Macromolecular Modelling, New York, October 12-14, 2000, T. Schlick and H. H. Gan, eds., Lecture Notes in Computational Science and Engineering, Vol. 24, pp. 1-25, Springer Verlag, Berlin (2002) -




J. Huang, Q. Zhang, and T. Schlick, "Effect of DNA Superhelicity and Bound Proteins on Mechanistic Aspects of the Hin-mediated and Fis-enhanced Inversion", Biophys. J., 85:804-817 (2003) 

-




J. Aishima, R. K. Gitti, J. E. Noah, H. H. Gan, T. Schlick, and C. Wolberger, "A Hoogsteen Base Pair Embedded in Undistorted B-DNA", Nuc. Acids Res., 30:5244-5252 (2002) -




H. H. Gan, S. Pasquali, and T. Schlick, "Exploring The Repertoire of RNA Secondary Motifs Using Graph Theory with Implications for RNA Design", Nuc. Acids Res., 31:2926-2943 (2003) 


-





D. Xie and T. Schlick, "A More Lenient Stopping Rule for Line Search Algorithms", Optim. Meth. Softw., 17:683-700 (2002) 

-





D. Xie, S. B. Singh, E. M. Fluder, and T. Schlick, "Principal Component Analysis Combined with Truncated-Newton Minimization for Dimensionality Reduction of Chemical Databases", Math. Program. Ser. B, 95:161-185 (2003) 


-





J. Huang and T. Schlick, "Macroscopic Modeling and Simulations of Supercoiled DNA with Bound Proteins", J. Chem. Phys., 117:8573-8586 (2002) 

-



D. Barash, T. Schlick, M. Israeli, and R. Kimmel, "Multiplicative Operator Splittings in Nonlinear Diffusion: From Spatial Splitting to Multiple Timesteps", J. Math. Imag. Vision, 19:33-48 (2003) 
-




D. Strahs, D. Barash, X. Qian, and T. Schlick, "Sequence-Dependent Solution Structure and Motions of 13 TATA/TBP Complexes", Biopolymers, 69:216-243 (2003) 



-





D. Barash, L. Yang, X. Qian, and T. Schlick, "Inherent Speedup Limitations in Multiple Timestep/ Particle Mesh Ewald Algorithms", J. Comp. Chem., 24:77-88 (2003) 

-




L. Yang, W. A. Beard, S. H. Wilson, B. Roux, S. Broyde, and T. Schlick, "Local Deformations Revealed by Dynamics Simulations of DNA Polymerase 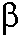 with DNA Mismatches at the Primer Terminus", J. Mol. Biol., 321:459-478 (2002)
with DNA Mismatches at the Primer Terminus", J. Mol. Biol., 321:459-478 (2002) 




-





X. Qian and T. Schlick, "Efficient Multiple Timestep Integrators with Distance-Based Force Splitting for Particle-Mesh-Ewald Molecular Dynamics Simulations", J. Chem. Phys., 116:5971-5983 (2002) 


-





P. F. Batcho and T. Schlick, "New Splitting Formulations for Lattice Summations", J. Chem. Phys., 115:8312-8326 (2001) 


-





L. Yang, W. A. Beard, S. H. Wilson, S. Broyde, and T. Schlick, "Polymerase 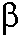 Simulations Reveal That Arg258 Rotation is a Slow Step Rather Than Large Subdomain Motions Per Se", J. Mol. Biol., 317:651-671 (2002)
Simulations Reveal That Arg258 Rotation is a Slow Step Rather Than Large Subdomain Motions Per Se", J. Mol. Biol., 317:651-671 (2002) 



-



T. Schlick, "Time-Trimming Tricks for Dynamic Simulations: Splitting Force Updates to Reduce Computational Work", Structure, 9:R45-R53 (2001) 


-





P. F. Batcho and T. Schlick, "Special Stability Advantages of Position Verlet Over Velocity Verlet in Multiple-Timestep Integration", J. Chem. Phys., 115:4019-4029 (2001) 


-





P. F. Batcho, D. A. Case, and T. Schlick, "Optimized Particle-Mesh Ewald/Multiple-Timestep Integration for Molecular Dynamics Simulations", J. Chem. Phys., 115:4003-4018 (2001) 


-





X. Qian, D. Strahs, and T. Schlick, "A New Program for Optimizing Periodic Boundary Models of Solvated Biomolecules (PBCAID)", J. Comp. Chem., 22:1843-1850 (2001) 


-





H. H. Gan, R. A. Perlow, S. Roy, J. Ko, M. Wu, J. Huang, S. Yan, A. Nicoletta, J. Vafai, D. Sun, L. Wang, J. E. Noah, S. Pasquali, and T. Schlick, "Analysis of Protein Sequence/Structure Similarity Relationships", Biophys. J., 83:2781-2791 (2002) 


-




X. Qian, D. Strahs, and T. Schlick, "Dynamic Simulations of 13 TATA Variants Refine Kinetic Hypotheses on Sequence/Activity Relationships", J. Mol. Biol., 308:681-703 (2001) 


-





H. H. Gan, A. Tropsha, and T. Schlick, "Lattice Protein Folding with Two and Four-Body Statistical Potentials", Proteins: Struc. Func. Gen., 43: 161-174 (2001) 


-




T. Schlick and L. Yang, "Long-Timestep Biomolecular Dynamics Simulations: LN Performance on a Polymerase Beta / DNA System", in Multiscale Computational Methods in Chemistry and Physics, Vol 177, NATO Science Series: Series III Computer and Systems Sciences, A. Brandt, J. Bernholc and K. Binder, eds., IOS Press, Amsterdam (2001) 


-






D. A. Beard and T. Schlick, "Computational Modeling Predicts the Structure and Dynamics of Chromatin Fiber", Structure, 9:105-114, February (2001) 


-





H. H. Gan, A. Tropsha, and T. Schlick, "Generating Folded Protein Structures with a Lattice Chain Growth Algorithm", J. Chem. Phys., 113:5511-5524, October (2000) 


-





J. Huang, T. Schlick, and A. Vologodskii, "Dynamics of Site Juxtaposition in Supercoiled DNA", Proc. Natl. Acad. Sci., 98:968-973, January (2001) 


-





T. Schlick, D. A. Beard, J. Huang, D. Strahs, and X. Qian, "Computational Challenges in Simulating Large DNA over Long Times", IEEE Computing in Science & Engineering, 2:38-51, Nov/Dec (2000) 


-




D. A. Beard and T. Schlick, "Modeling Salt-Mediated Electrostatics of Macromolecules: The Discrete Surface Charge Optimization Algorithm and Its Application to the Nucleosome", Biopolymers, 58:106-115 , January (2001) 


-






D. Strahs and T. Schlick, "A-tract Bending: Insights into Experimental Structures by Computational Models", J. Mol. Biol., 301:643-663, August (2000) 


-




D. A. Beard and T. Schlick, "Inertial Stochastic Dynamics. I. Long-time-step Methods for Langevin Dynamics", J. Chem. Phys., 112:7313-7322, May (2000) 


-




D. A. Beard and T. Schlick, "Inertial Stochastic Dynamics. II. Influence of Inertia on Slow Kinetic Processes of Supercoiled DNA", J. Chem. Phys., 112:7323-7338, May (2000) 


-





D. Xie and T. Schlick, "Visualization of Chemical Databases Using the Singular Value Decomposition and Truncated-Newton Minimization", Optimization in Computational Chemistry and Molecular Biology: Local and Global Approaches, Volume 40, pages 267-286, C. A. Floudas and P. M. Pardalos, eds., Kluwer Academic Publishers, Dororecht/Boston/London (2000) 


-



T. Schlick, "Computational Molecular Biophysics Today: A Confluence of Methological Advances and Complex Biomolecular Applications", J. Comp. Phys., 151:1-8, May (1999) 
-




A. Sandu and T. Schlick, "Resonance Analysis in Force Splitting Methods for Biomolecular Dynamics", J. Comp. Phys., 151:74-113, May (1999) 
-




T. Schlick, R. D. Skeel, A. T. Brunger, L. V. Kalé, J. Hermans, K. Schulten, and J. A. Board, Jr., "Algorithmic Challenges in Computational Molecular Biophysics", J. Comp. Phys., 151:9-48, May (1999) 

-




D. Xie and T. Schlick, "Efficient Implementation of the Truncated Newton Method for Large Scale Chemistry Applications", SIAM J. Opt., 10: 132-154 October (1999) 


-




D. Xie and T. Schlick, "Remark on the Updated Truncated Newton Minimization Package, Algorithm 702", ACM Trans. Math. Softw., 25, 108-122, March (1999) 


-




D. Xie, L. R. Scott and T. Schlick, "Analysis of the SHAKE-SOR Algorithm for Constrained Molecular Dynamics Simulations", Methods and Applications of Analysis, a Special issue dedicated to Prof. Cathleen Morawetz of the Courant Institute, 7: 577-590, September (2000) 


-




D. Xie, A. Tropsha and T. Schlick, "An Efficient Projection Protocol for Chemical Databases: Singular Value Decomposition Combined with Truncated-Newton Minimization", J. Chem. Inf. Comput. Sci., 40:167-177, December (1999) 


-




E. Barth and T. Schlick, "I. Overcoming Stability Limitations in Biomolecular Dynamics: Combining Force Splitting via Extrapolation with Langevin Dynamics in LN", J. Chem. Phys., 109:1617-1632, August (1998) 


-




E. Barth and T. Schlick, "II. Extrapolation Versus Impulse in Multiple-Timestepping Schemes: Linear Analysis and Applications to Newtonian and Langevin Dynamics", J. Chem. Phys. 109:1633-1642, August (1998) 


-




P. Derreumaux and T. Schlick, "Simulation of the Loop Opening/Closing of the Enzyme Triosephosphate Isomeraze (TIM)", Biophys. J., 74:72-81, January (1998) 


-




H. Jian, T. Schlick, and A. Vologodskii, "Internal Motion of Supercoiled DNA: Brownian Dynamics Simulations of Site Juxtaposition", J. Mol. Biol., 284:287-296, November (1998) 


-




T. Schlick, "Geometry Optimization", Contributed chapter to the Encyclopedia of Computational Chemistry (5 Volumes), Volume 2, pages 1136-1157, P. von Rague Schleyer, Editor in Chief and N. L. Allinger, T. Clark, J. Gasteiger, P. A. Kollman, and H. F. Schaefer III, eds., John Wiley & Sons, West Sussex (1998) 
-



T. Schlick, "Some Failures and Successes of Long-Timestep Approaches to Biomolecular Simulations", in Algorithms for Macromolecular Modeling, Volume 4, pages 227-262, Springer-Verlag series in "Lecture Notes in Computational Science and Engineering", P. Deuflhard, J. Hermans, B. Leimkuhler, A. Mark, R. D. Skeel, and S. Reich, eds. (1998) 


-




T. Schlick, M. Mandziuk, R. D. Skeel, and K. Srinivas, "Nonlinear Resonance Artifacts in Molecular Dynamics", J. Comp. Phys., 139:1-29, February (1998) 


-



L. Y. Zaslavsky and T. Schlick, "An Adaptive Multigrid Technique for Evaluating Long-Range Forces in Biomolecular Simulations", App. Math. Comp., 97:237-250, December (1998) 


-



E. Barth, M. Mandziuk, and T. Schlick, "A Separating Framework for Increasing the Timestep in Molecular Dynamics", contributed chapter to Computer Simulation of Biomolecular Systems, Volume 3, Chapter 4, pages 97-120, W. van Gunsteren, P. Weiner and T. Wilkinson eds., ESCOM Science Publishers, Ledien, The Netherlands (1997) 


-




H. Jian, A. Vologodskii, and T. Schlick, "A Combined Wormlike Chain and Bead Model for Dynamic Simulations of Long DNA", J. Comp. Phys., 136:168-179, September (1997) 

-



G. Liu, T. Schlick, A. J. Olson, and W. K. Olson, "Configurational Transitions in DNA Supercoils Represented by Fourier-Series Curve-Fitting Techniques", Biophys. J., 73:1742-1762, October (1997) 

-




G. Ramachandran and T. Schlick, "On Buckling Transitions in Supercoiled DNA: Dependence on Elastic Constants and DNA Size", Biopolymers, 41:5-25, January (1997) 


-




T. Schlick, E. Barth, and M. Mandziuk, "Biomolecular Dynamics at Long Timesteps: Bridging the Time Scale Gap Between Simulation and Experimentation", Annu. Rev. Biophys. Biomol. Struct., 26:181-222 (1997) 


-




R. D. Skeel, G. Zhang and T. Schlick, "A Family of Symplectic Integrators: Stability, Accuracy, and Molecular Dynamics Applications", SIAM J. Sci. Comp., 18:203-222 (1997) 


-




B. Mishra and T. Schlick, " The Notion of Error in Langevin Dynamics: (1) Linear Analysis", J. Chem. Phys., 105:299-318, July (1996) 


-




T. Pinou, T. Schlick, B. Li, and H. Dowling, "Addition of Darwin's Third Dimension to Evolutionary Trees", J. Theor. Bio., 219:505-512, October (1996) 

-



G. Ramachandran and T. Schlick, "Beyond Optimization: Simulating the Dynamics of Supercoiled DNA by a Macroscopic Model", in DIMACS Series in Discrete Mathematics and Theoretical Computer Science, Volume 23, Global Minimization of Nonconvex Energy Functions: Molecular Conformation and Protein Folding, P. Pardalos, D. Shalloway, and G. Xue, eds., pages 215-231, American Mathematical Society, Providence, Rhode Island (1996) 

-



T. Schlick, "Pursuing Laplace's Vison on Modern Computers", Proceedings of the IMA Program in Mathematical Biology, IMA Volumes in Mathematics and its Applications, Volume 82, pages 219-247, J. Mesirov, K Schulten, and D. W. Sumners, eds., Springer-Verlag, New York (1996) 


-



T. Schlick and A. Brandt, "A Multigrid Tutorial with Applications to Molecular Dynamics", IEEE Comp. Sci. Eng., 3:78-83, Fall (1996) 
-



P. Derreumaux and T. Schlick, "Long Timestep Dynamics of Peptides by the Dynamics Driver Approach", Proteins: Struc. Func., Gen., 21:282-302, April (1995) 


-



G. Liu, W. K. Olson, and T. Schlick, "Application of Fourier Analysis to Computer Simulation of Supercoiled DNA Formation", Comp. Polymer Sci., 5:7-27 (1995) 


-




M. Mandziuk and T. Schlick, "Resonance in the Dynamics of Chemical Systems Simulated by the Implicit Midpoint Scheme", Chem. Phys. Lett., 237:525-535, March (1995) 


-




G. Ramachandran and T. Schlick, "Solvent Effects on Supercoiled DNA Explored by Langevin Dynamics Simulations", Phys., Rev. E., 51:6188-6203, June (1995) 



-



T. Schlick, "Modeling Superhelical DNA: Recent Analytical and Dynamic Approaches", Curr. Opin. Struc. Bio., 5:245-262, April (1995). Special issue on Theory and Simulation, B. Honig, ed. 


-



T. Schlick and C. S. Peskin, "Comment on 'Backward Euler and other methods for simulating molecular fluids'", J. Chem. Phys., 103:9888-9889, December (1995) 
-



G. Zhang and T. Schlick, "Implicit Integration Schemes for Langevin Dynamics", Mol. Phys. 84:1077-1098 (1995) 



-



V. Z. Averbukh, S. Figueroa, and T. Schlick, "Remark on Algorithm 566", ACM Trans. Math. Softw., 20:282-285, September (1994) -



J. A. Board, Jr., L. V. Kalé, K. Schulten, R. D. Skeel, and T. Schlick, "Modeling Biomolecules: Larger Scales, Longer Durations", IEEE Comp. Sci. Eng., 1:19-30, Winter (1994) 
-



P. Derreumaux, G. Zhang, B. Brooks, and T. Schlick, "A Truncated-Newton Method Adapted for CHARMM and Biomolecular Applications", J. Comp. Chem., 15:532-552, April (1994) 

-



T. Schlick, B. Li, and M. H. Hao, "Calibration of the Timestep for Molecular Dynamics of Supercoiled DNA Modeled by B-Splines", in Structural Biology, State of the Art 1993, Volume 1, Proceedings of the Eighth Conversation, M. H. Sarma and R. S. Sarma, eds., pages 157-174, Adenine Press, Guiderland, New York (1994) 


-



T. Schlick, B. Li, and W. K. Olson, "The Influence of Salt on the Structure and Energetics of Supercoiled DNA", Biophys. J., 67:2146-2166, December (1994) 



-



T. Schlick, W. K. Olson, T. Westcott, and J. P. Greenberg, "On Higher Buckling Transition in Supercoiled DNA", Biopolymers, 34:565-597, May (1994) 



-



G. Zhang and T. Schlick, "The Langevin/Implicit-Euler/Normal-Mode Scheme (LIN) for Molecular Dynamics at Large Timesteps", J. Chem. Phys., 101:4995-5012, September (1994) 



-



T. Schlick, "Modified Cholesky Factorizations for Sparse Preconditioners", SIAM J. Sci. Comp., 14:424-445, March (1993) 

-



T. Schlick, W. K. Olson. T. Westcott, G. Liu, and J. P. Greenberg, "Supercoiled Configurations of Small DNA Rings and Their Dynamics", Abstract of the Eigth Conversation in Biomolecular Stereodynamics, J. Biomol. Struct. Dyn., 10 a170 (1993) -



G. Zhang and T. Schlick, "LIN: A New Algorithm Combining Implicit Integration and Normal Mode Techniques for Molecular Dynamics", J. Comp. Chem., 14:1212-1233, July (1993) 
-



X. Zou, I. M. Navon, M. Berger, P. K. H. Phua, T. Schlick and F. X. Le Dimet, "Numerical Experience with Limited-Memory and Truncated Newton Methods", SIAM J. Opt., 3:582-608, August (1993) 

-



V. Z. Averbukh, S. Figueroa, and T. Schlick, "HESFCN -A FORTRAN Package of Hessian Subroutines for Testing Nonlinear Optimization Software", Computer Science Technical Report, 610:1-43, June (1992) -



A. Nyberg and T. Schlick,"On Increasing the Time Step in Molecular Dynamics", Chem. Phys. Lett., 198:538-546, October (1992) 
-



T. Schlick, "Optimization Methods in Computational Chemistry", in Reviews in Computational Chemistry, Volume 3 , Chapter 1, pages 1-71, K. B. Lipkowitz and D. B. Boyd, eds., VCH Publishers, New York (1992) 



-



T. Schlick and A. Fogelson, "TNPACK - A Truncated Newton Minimization Package for Large Scale Problems: I. Algorithm and Usage", ACM Trans. Math. Softw., 18:46-70, March (1992) 


-



"T. Schlick and A. Fogelson, "TNPACK - A Truncated Newton Minimization Package for Large Scale Problems: II. Implementation Examples", ACM Trans. Math. Softw., 18:71-111, March (1992) 


-



T. Schlick and W. K. Olson, "Computer Simulations of Supercoiled DNA Energetics and Dynamics", J. Mol. Biol., 223:1089-1119, February (1992) 

-



T. Schlick and W. K. Olson, "Trefoil Knotting Revealed by Molecular Dynamics of Supercoiled DNA", Science, 257:1110-1115, August (1992) 



-



A. Nyberg and T. Schlick, "A Computational Investigation of Dynamic Properties with the Implicit-Euler Scheme for Molecular Dynamics Situations", J. Chem. Phys., 95:4986-4996, October (1991) -



T. Schlick, "New Approaches to Potential Energy Minimization and Molecular Dynamics Algorithms", based on an invited presentation at the Molecular Mechanics and Molecular Dynamics Workshop , Florida State University, Tallahassee, Comput. Chem., 15:251-260 (1991) 

-



T. Schlick, S. Figueroa, and M. Mezei, "A Molecular Dynamics Simulation of a Water Droplet by the Implicit-Euler/Langevin Scheme", J. Chem. Phys., 94:2118-2129, February (1991) 

-



T. Schlick, B. E. Hingerty, C. S. Peskin, M. L. Overton, and S. Broyde, "Search Strategies, Minimization Algorithms, and Molecular Dynamics Simulations for Exploring Conformational Spaces of Nucleic Acids", in Theoretical Biochenmistry and Molecular Biophysics, Volume 1, pages 39-58, D. L. Beveridge and R. Lavery, eds., Adenine Press, Guilderland, New York (1991) 

-



T. Schlick, "Implementation of the Schnabel and Eskow Modified Cholesky Factorization in the Context of Large-Scale Nonlinear Optimization", Computer Science Technical Report, Courant Institute 525:1-40, (1990) 

-



C. S. Peskin and T. Schlick, "Molecular Dynamics by the Backwards-Euler Method", Comm. Pure Appl. Math., 42:1001-1031 (1989) 
-



T. Schlick, "A Recipe for Evaluating and Differentiating Cos  Expressions", J. Comp. Chem., 10:951-956, May (1989)
Expressions", J. Comp. Chem., 10:951-956, May (1989) -



T. Schlick and C. S. Peskin, "Can Classical Equations Simulate Quantum Mechanical Behavior? A Molecular Dynamics Investigation of a Diatomic Molecule with a Morse Potential", Comm. Pure Appl. Math., 42:1141-1163 (1989) 
-



T. Schlick, C. S. Peskin, and M. L. Overton, "A New Pair of Algorithms for Potential Energy Minimization and Molecular Dynamics Simulations", Book of Abstracts of the Sixth Conversation in Biomolecular Stereodynamics, Volume 151, pages 74-113, R. H. Sarma, ed., State University of New York, Albany (1989) -


T. Schlick, "A Modular Strategy for Generating Starting Conformations and Data Structures of Polynucleotide Helices for Potential Energy Calculations", J. Comp. Chem., 9:861-889, June (1988) 
-



T. Schlick, "Modeling and Minimization Techniques for Predicting Three-Dimensional Structures of Large Biological Molecules", Ph. D. Dissertation, Courant Institute, Department of Mathematics, New York University (1987). -



T. Schlick and M. Overton, "A Powerful Truncated Newton Method for Potential Energy Minimization", J. Comp. Chem., 8:1025-1039, May (1987) 

-



T. Schlick, C. Peskin, S. Broyde, and M. Overton, "An Analysis of the Structural and Energetic Properties of Deoxyribose by Potential Energy Methods" J. Comp. Chem., 8:1199-1224, June (1987) 

-



T. Schlick, C. Peskin, S. Broyde and M. Overton, "Development of a New Computational Approach for the Prediction of Nucleic Acid Structure by Potential Energy Methods: I. Deoxyribose", Computer Science Technical Report, Courant Institute, 264:1-60 (1986) 
-



W. L. Hase, D. M. Ludlow, R. J. Wolf and T. Schlick, "Translational and Vibrational Energy Dependence of the Cross Section for H + C2H4 ---> C2H5" J. Phys. Chem., 85:958-968 (1981)