|
Chemistry: G25.2601
Biology: G23.2601
Mathematics: G63.2856.003
Computer Science: G22.3033.11
Sackler: G16.2607
Time: Thursdays, 12:45-2:45pm
Location: 1003 Main Building
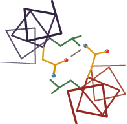
Kinemage (``Kinetic Images'') Tutorial
Tamar Schlick
The kinemage program created by Dave and Jane Richardson of Duke University
is a wonderful way to study protein structure. Their program offers
instructive sets of biomolecular images that can be manipulated
in various ways so as to understand the three-dimensional complexity
and variability of biomolecular systems.
Below are instructions on how to use the kinemage tutorial. Basically,
you will need to first
download two types of files: the driver program that manipulates kinemage
files and the data files themselves. The driver program can be taken for
Mac, PC, SGI machines, and other computers. The instructions below refer to
SGI platforms, but you will simply need to transfer a different driver file for
another machine; file names are self-explanatory.
You can obtain some general information on kinemage through the Richardsons'
web site: kinemage.biochem.duke.edu.
The instructions below assume you have basic knowledge of transferring
files via ftp. If you do not, read the online manual (type: man ftp).
To run the kinemage programs using the SGI driver, you will need an
SGI machine with SGI operating system version 6.2 or higher.
-
Transfer MAGE and kinemage files
-
FTP to the resource site by typing at the Unix command line
ftp://kinemage.biomchem.duke.edu
Log in as anonymous. You will now be receiving ftp prompts as:
ftp>
- Go to the appropriate subdirectory by typing at the ftp command line:
cd UNIXprograms
- Specify binary mode of transfer on the ftp command line as follows:
binary
(The file you transfer is an executable file, so an ASCII (text) transfer mode
will cause error when you want to run the program).
- Now get the kinemage driver program:
get MAGE_5.30_990119.SGI
(To ensure correct file transfer, it is a good idea to compare
the size of the retrieved file with that of the original).
- Now scroll to a different subdirectory
where the kinemage input files reside:
cd ../KINfiles
- Check the list of available files by typing:
ls
You should see several files, including
ProTour1.kin, ProTour2.kin, ProTour3.kin, ProTour4.kin, ProTour5.kin,
ProTour6.kin, ProTour7.kin, ProTour9.kin, and NATour1.kin.
- Retrieve each of the above files in turn by using the ftp command:
get
where
represents one of the above titles. Apparently,
the remote server does not support the multiple retrieving mode
at the time of this writing.
- Exitfrom from ftp by typing:
quit
Check that you have all the necessary files by typing the Unix commands
ls *.kin
and then
ls MAGE*
-
Run the kinemage program
- In your Unix command line, change the mode of the file
MAGE_5.30_990119.SGI to executable by typing:
chmod +x MAGE_5.30_990119.SGI
- Start the program by tying:
MAGE_5.30_990119.SGI
You will see four windows, with the front window
(of title MAGE GRAPHICS) awaiting your response.
Press the proceed button to hide the front window. The
two smaller windows may be behind the main one;
try move the main window aside so that the others,
with titles MAGE TEXT and MAGE CAPTION,
will be in sight.
- To use kinemages to study protein structures, pull
down the main window's file menu, and choose the open new file...
to start a session. You can select (or type in the name) of one of your transferred
.kin files, and then press the OK button.
The main window will display a
picture of a biomolecule,
while the MAGE TEXT window will provide explanatory text
about the system. The MAGE CAPTION window shows
the information of the current kinemage in view.
There are typically several kinemages in the same .kin file.
- To view another kinemage in the same file, select Next
or Choose from the
Kinemage menu.
- To manipulate the image, rotate it by moving the mouse with
the left button pressed. You can also click on the boxes on the
right side to turn off/on various displays or markers.
Read the help text by clicking on the Help Mage menu and choosing a topic.
Content of MAGE Files
Below is a brief synopsis of the tutorial .kin files for easy
reference; descriptions
are taken straight from MAGE TEXT. I suggest the following order of study.
Protein Classification and Motifs
- ProTour7.kin: Major Protein Folding Motifs
(
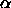 , , 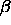 , , 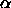 / /  ) )
Each of these kinemages shows ribbon-diagrams (one-strand, smooth
splines along the Calpha backbone) for a sample of protein structures
typical of one of the major categories of tertiary structure. In each
kinemage, click on the ``ANIMATE" button to switch among the example
proteins. The ribbons are depth-cued by line width, secondary-structure
features are color-coded, prosthetic groups are shown, and the N- and
C-termini are labeled. Clicking on a point along the ribbon will display
the residue name and number at the bottom left of the screen. Drag slowly
with the mouse to rotate the molecule, for a 3-D perception, or hit the
``s" key on the keyboard to toggle side-by-side stereo on and off. (NOTE:
use the ANIMATE button to switch among these examples since this invokes
special recentered views.)
Kin.1- All-Alpha structures: myohemerythrin, cytochrome b562, & calbindin
Kin.2- Parallel Alpha/Beta structures: flavodoxin & triose P isomerase
Kin.3- Antiparallel Beta structures: Strep. subtilisin inhibitor, trypsin d2, & STNV
Kin.4- Small Irregular structures: BPTI, crambin, & cytochrome c3 -
-
ProTour1.kin:
 Sheet Motif Sheet Motif
The doubly-wound parallel beta sheet, a subset of the Parallel
Alpha/Beta type of tertiary structure, is the commonest folding pattern
found in the known protein structures. This ``fold" is also known as the
``nucleotide-binding domain", because most of them bind a mononucleotide
(such as FMN) or a dinucleotide (such as NAD) near the middle of one end
of the beta sheet. This month's Protein Tourist illustrates the main
features of this protein ``fold", to be used for general information
purposes or for teaching. No further permission is needed for educational
use of kinemages from Protein Tourist or Demo files.
Kin.1 - A parallel beta sheet, with H-bonds
Kin.2 - Right-handedness of crossover connections
Kin.3 - LDH domain 1, with animation of the ``double winding"
Kin.4 - ADH domain 2, another classic doubly-wound sheet
Kin.5 - DHFR, a less regular doubly-wound sheet
Kin.6 - Nucleotide binding in LDH, with helix dipole and moving loop
Kin.7 - Domain organization in LDH
- ProTour2.kin: Leucine Zipper
A motif known as the ``leucine zipper" mediates dimerization of the
bZIP and bHLH-ZIP classes of transcription factors. Leu zipper sequences are
characterized by a sequence of 30 or 40 helix-tolerating amino acids with
Leu every seven residues. Leucine zipper peptides form dimers of parallel
helices proposed to be similar to coiled coils common in much longer
alpha-fibrous proteins. A recent high-resolution
x-ray structure of a synthetic, 33-residue fragment of the yeast
transcription factor GCN4 confirms that the leucine zipper forms a coiled
coil, and illustrates interactions that mediate dimerization. This Protein
Tourist allows readers to explore the GCN4 leucine zipper structure.
Kin.1 - Tropomyosin Calphas: a long coiled-coil in a fibrous protein
Kin.2 - A short, distorted coiled coil in the dimer interface of CAP protein
Kin.3 - GCN4 leucine zipper peptide: a high-resolution coiled-coil
structure
Kin.4 - Salt bridges around the outside of the zipper
hydrophobic core
Kin.5 - Asymmetrical arrangement of Asn 16
- ProTour4.kin: Disuphide-Bond-Rich Proteins
Small SS-rich, C-centered overhand structures:
Kin.1 - Cellobiohydrolase C-terminal domain: overhand topology and C-C beta
hairpin
Kin.2 - Potato carboxypeptidase inhibitor: very small SS-rich
Kin.3 - Scorpion neurotoxin
Kin.4 - Closeup of glycine roles, in scorpion toxin
Small SS-rich, N-centered overhands and others:
Kin.5 - Ovomucoid serine protease inhibitor: N-centered topology
Kin.6 - Wheat germ agglutinin domain
Kin.7 - Hirudin: up & down topology, with 3-SS core
Small metal-rich structures
Kin.8 - Rubredoxin: one non-heme Fe
Kin.9 - Ferredoxin (P. aer.): two Fe4S4 clusters
Kin.10- Cytochrome C3: 4 hemes
Kin.11- Metallothionein domain 2: 4 Cd sites in 31 residues
Small proteins with neither disulfides nor metals
Kin.12- Immunoglobulin-binding domain of Strep. protein G
Specific Proteins
- ProTour3.kin:
 Repressor Repressor
Kin.1 - Overview, with Lambda repressor Calphas and virtual-bond
DNA
Kin.2 - DNA, for consensus half-operator sequence, with bases
color-coded
Kin.3 - Detail of sequence-specific interactions with bases in
one major groove
Kin.4 - Non-specific interactions with phosphate backbone
- ProTour5.kin: Carboxypeptidase
Carboxypeptidase:
Kin.1 - Overview of the 3-D structure of
carboxypeptidase A
Kin.2 - Carboxypeptidase active site motions on substrate binding
Kin.3 - Structure of a tetrahedral transition-state
analog at the active site, animated
Kin.4 - Potato carboxypeptidase inhibitor, as bound to CPA
Thermolysin
Kin.5 - Thermolysin: overall structure
Kin.6 -
Closeup of ZFpLA bound at thermolysin active site
Kin.7 - Comparison of
thermolysin and carboxypeptidase, Calphas & active sites
- ProTour6.kin: Inhibitor Complexes
Kin.1 - Thrombin, with backbone, Ser-His-Asp-Ser, PPACK inhibitor, and
beta barrels
Kin.2 - Trypsin-BPTI complex: overview of inhibitor binding
Kin.3 - Trypsin-BPTI complex: detail of active site, with small-probe dot
contacts
Kin.4 - Superposition of trypsin, chymotrypsin, and subtilisin
- ProTour9.kin: Pectate Lyase C
Pectate Lyase C (PelC) has a novel 3-dimensional structure, made
up almost entirely of an L-shaped, or triangular, coil of parallel beta
sheet. This represents a new major category of protein
tertiary-structure, the all-beta parallel category.
Kin.1 - Pectate Lyase C: buildup of the parallel beta coil, in sequence order
Kin.2 - PelC: an animated tour of features, on the Calpha backbone
Kin.3 - Sidechain stacking in the interior of PelC: the Asn ladder
DNA
- NATour1.kin: B-DNA Oligomers
Kin.1 - 5'-D( C G C G A A T T C G C G)-3', 290 (NDB file: BDL001)
Kin.2 - 5'-D( C G C A T A T A T G C G)-3' (BDL007)
Kin.3 - 5'-D( C G C A A G C T G G C G)-3' (BDL022)
Kin.4 - 5'-D( C G C G (M)A A T T C G C G)-3' (BDLB13)
Kin.5 - 5'-D( C G C G A A T T (BR)C G C G)-3', NETRO SIN (GDLB05)
Kin.6 - 5'-D( C G C A A A T T T G C G)-3', DISTAMYCIN (GDL003)
Kin.7 - 5'-D( C G C G A A T T C G C G)-3', BERENIL (GDL009)
Kin.8 - 5'-D( C G C G A A T T C G C G)-3',4'-6-DIAMIDINE-2-PHENYL
INDOLE (DAPI) (GDL008)
For further information, contact T. Schlick by email (schlick@nyu.edu)
phone (998-3116) or fax (995-4152).
|