Introduction
Many recent discoveries on how biological and synthetic RNAs can perform a broad range of functions are changing our traditional view ("central dogma" in Figure 1) that the primary role of RNA is to mediate the synthesis of proteins, the cell's workhorses. Indeed, we well appreciate that many fundamental cellular processes such as gene regulation and enzymatic catalysis are performed by RNA. For example, small regulatory RNAs (e.g., miRNAs, siRNA) can modulate the expression of many proteins and RNA enzymes (ribozymes, Figure 2) can catalyze a variety of chemical reactions. Significantly, the discovery that thousands mammalian RNA transcripts do not code for proteins may indicate a far greater role for cellular RNAs than previously imagined. In addition to biological RNAs, synthetic RNAs developed from in vitro selection – an experimental technique for identifying active RNAs from random sequence pools – have led to many ligand-binding and catalytic RNAs. These advances in RNA highlight the versatility of RNA molecules. Emerging applications of engineered or designed RNAs include RNA nanotechnology, where RNAs are assembled into functional arrays, and RNA synthetic biology, where designed RNAs are used to control cellular functions (e.g., regulate gene expression). These exciting advances offer new investigative and application tools for molecular biology, proteomics, and molecular medicine.
Our modeling of RNA structure and function using graphs with our RNA-As-Graphs (RAG) framework aims to discover novel active RNA molecules via analysis, folding and design of RNA sequences and structures. In RNA analysis, our group has contributed to RNA graphical representations to describe RNA’s structural repertoire, applied bioinformatics tools to identify RNA’s tertiary interaction motifs, and developed tools for analyzing large sets of putative noncoding RNAs. Currently, we are focusing on new therapeutic strategies for viral infections by graph-based “inverse folding tools”.
Since the Covid-19 pandemic, we have applied our RNAs-As-Graphs modeling strategies to study the SARS-CoV-2 frameshifting element (FSE). Our work has uncovered a complex conformational landscape of pseudoknot and non-pseudoknot forms, developed minimal mutations of the FSE to transform the FSE into other folds or suppress structural transitions, and developed new energy-landscape interrogatory tools combining evolution and biophysics to study FSE motifs changes and develop innovative therapeutic strategies (Figure 3).
Figure 1. Central dogma of molecular biology
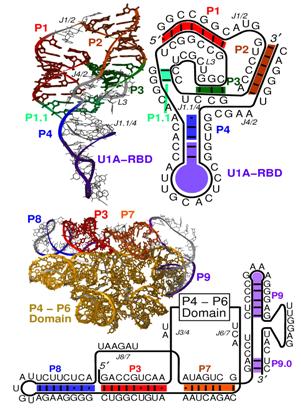
Figure 2. HDV ribozyme (top) and group I intron (bottom) from T. Schlick, “Molecular Modeling and Simulation: an Interdisciplinary Guide”, New York, Springer (2002)
Figure 3. Evolution of coronavirus FSE studied via RAG framework. Using graph theory-based methods for representing RNA secondary structures, conformational landscapes of viral FSEs are calculated with increasing sequence lengths for 23 representative Alpha and Beta-coronaviruses to investigate sequence influence and length dependence on virus structural evolution.
Recent highlights:
See our RAG Tutorial here