The genome contains the master blueprint for an organism encoded onto strands of DNA. In order for an organism to survive, DNA must be accurately replicated and damage repaired. Damage to the genome can occur from a number of exogenous and endogenous causes. It is estimated that the DNA in every cell of the human body is spontaneously damaged more than 10,000 times every day. To circumvent this problem, cells have evolved sophisticated machinery to replicate and repair DNA accurately and efficiently.
DNA polymerases orchestrate the addition of new nucleotides to a growing chain of DNA by catalyzing a nucleotidyl-transfer reaction for the incorporation of nucleotides one at a time. The relative ability of a polymerase to incorporate a correct nucleotide rather than an incorrect unit from a pool of structurally similar molecules is a measure of fidelity. When an incorrect nucleotide (e.g., G opposite A instead of T) is inserted, the blueprint directing the cell is changed. Since the accumulation of such errors is thought to play a key role in various disease states such as cancer and even the aging process, understanding polymerase mechanisms at the atomic level is very important.
DNA polymerases are found in primitive viruses and bacteria to complex multi-cellular organisms, including human beings. Based on sequence and structural similarities, seven different DNA polymerase families have been identified. Because they are crucial for maintaining genomic integrity, these polymerase families are evolutionarily conserved, and members are often found in a wide range of organisms.
Our work focuses on a representative group of repair DNA polymerases that includes X-family DNA polymerases β (pol β), X (pol X), λ (pol λ), and μ (pol μ); and Y-family DNA polymerase IV (Dpo4). Unlike high fidelity DNA replication polymerases, these lower fidelity enzymes lack an editing domain and typically have specialized functions which require limited nucleotide insertion. Some of these polymerases are involved in the repair of small oxidative lesions and double strand breaks while others specialize in bulky lesion bypass.
Each polymerase has a characteristic fidelity and error specificity. Our research focuses on their structure-function relationships and the various mechanisms that ensure replication fidelity. Although many high resolution X-ray crystal structures are available for these and other DNA polymerases, they only provide a static snapshot of the enzyme while interpreting how polymerases work often requires a dynamic picture. Furthermore, available experimental kinetic data provide information on polymerase behavior on the macroscopic level (such as fidelity and efficiency rates, error characteristics, etc.), but often insights are required at the atomic level.
Computer simulations can help bridge the gap between X-ray structures and observed characteristics by providing critical information on how polymerases operate at the atomic level. While subject to certain well-known limitations such as imperfect force fields and limited sampling, molecular dynamics (MD) simulations have proven capable of providing useful data on the motions of biological complexes. Going beyond X-ray structures, MD can show, for example, how polymerases bind to a substrate. However, some reactions involving small molecules like nucleotides are substantially affected by quantum mechanical forces and simple MD simulations are not sufficient to study them accurately. Due to computational limitations, such cases are dealt with in a split calculation. The chemical reaction is simulated for highest accuracy using quantum mechanics and the remaining environment is modeled using classical MM. This mixed QM/MM representation has proven quite useful in studying both details of the catalytic reaction and the effect of the remaining protein, DNA, and solvent environment (Figure 1). Our polymerase work also applied enhanced sampling methods to study complete opening/closing pathways (Figure 2).
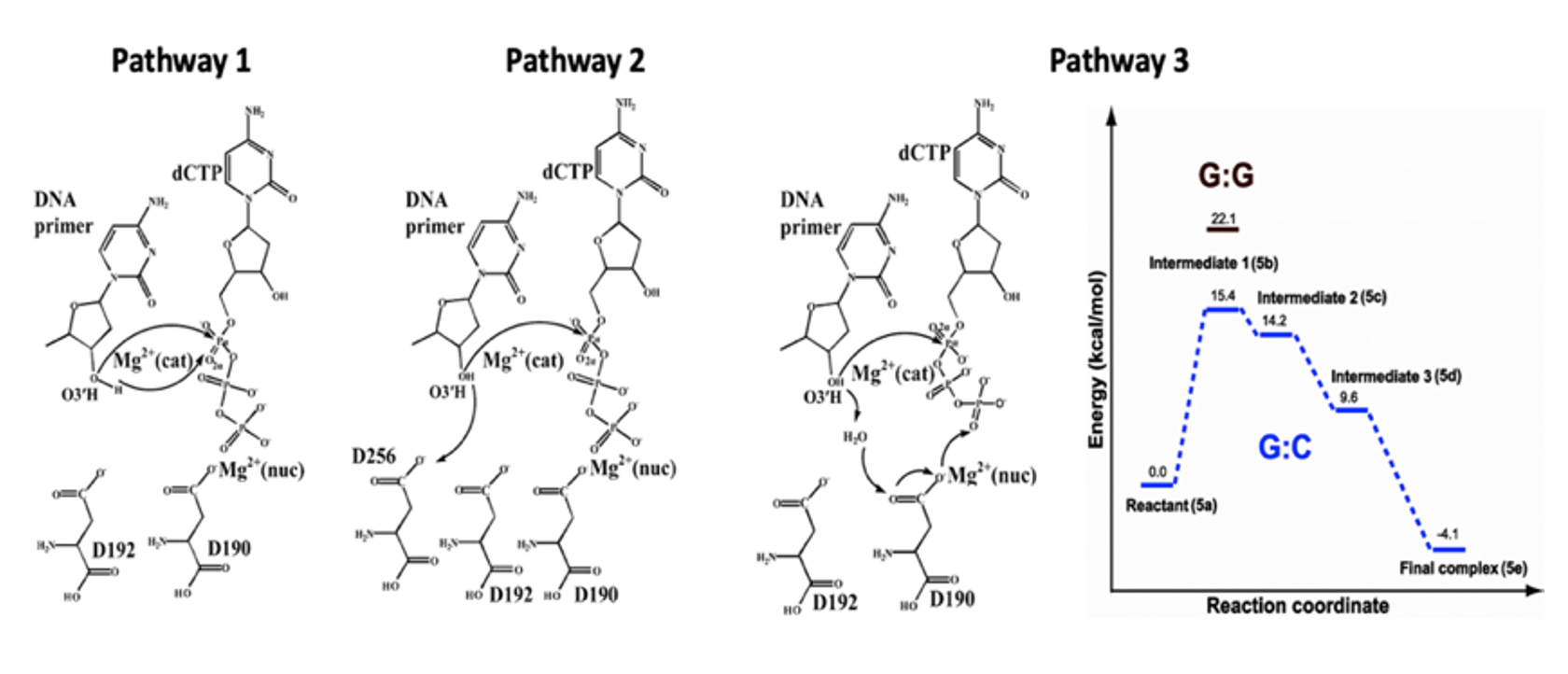
Figure 1. Explored reaction pathways for G incorporation by DNA pol beta determined with QM/MM simulations.

Figure 2. Fingers closing mechanism proposed by TPS simulations.
Research highlights:
• Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide
• Orchestration of cooperative events in DNA synthesis and repair mechanism unraveled by transition path sampling of DNA polymerase β's closing
• Critical Role of Magnesium Ions in DNA Polymerase β's Closing and Active Site Assembly
• Biomolecular Free Energy Profiles by a Shooting/Umbrella Sampling Protocol, `BOLAS'
• DNA Polymerase β Catalysis: Are Different Mechanisms Possible?